Vectors for Gene cloning Plasmids Bacteriophages khadijahhanimunimap edu

Vectors for Gene cloning: Plasmids & Bacteriophages khadijahhanim@unimap. edu. my Week 10: 21&22 April 2014

Course Outcome • Able to employ techniques used in the isolation, manipulation and cloning of heterologous DNA. VEHICLES FOR GENE CLONING Demonstrate the basic features of plasmids and bacteriophages and analyze their mechanism as cloning vehicles.
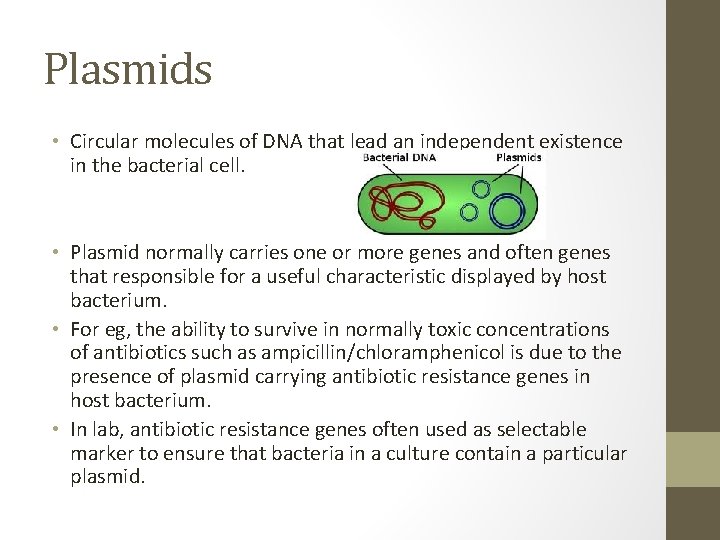
Plasmids • Circular molecules of DNA that lead an independent existence in the bacterial cell. • Plasmid normally carries one or more genes and often genes that responsible for a useful characteristic displayed by host bacterium. • For eg, the ability to survive in normally toxic concentrations of antibiotics such as ampicillin/chloramphenicol is due to the presence of plasmid carrying antibiotic resistance genes in host bacterium. • In lab, antibiotic resistance genes often used as selectable marker to ensure that bacteria in a culture contain a particular plasmid.
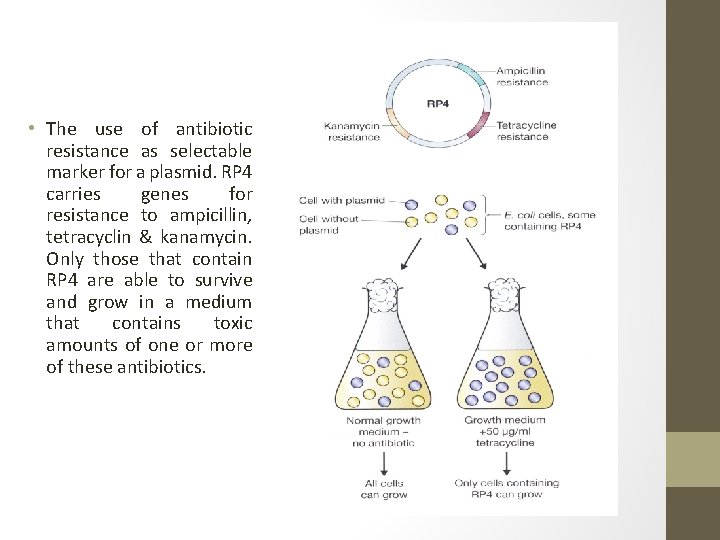
• The use of antibiotic resistance as selectable marker for a plasmid. RP 4 carries genes for resistance to ampicillin, tetracyclin & kanamycin. Only those that contain RP 4 are able to survive and grow in a medium that contains toxic amounts of one or more of these antibiotics.
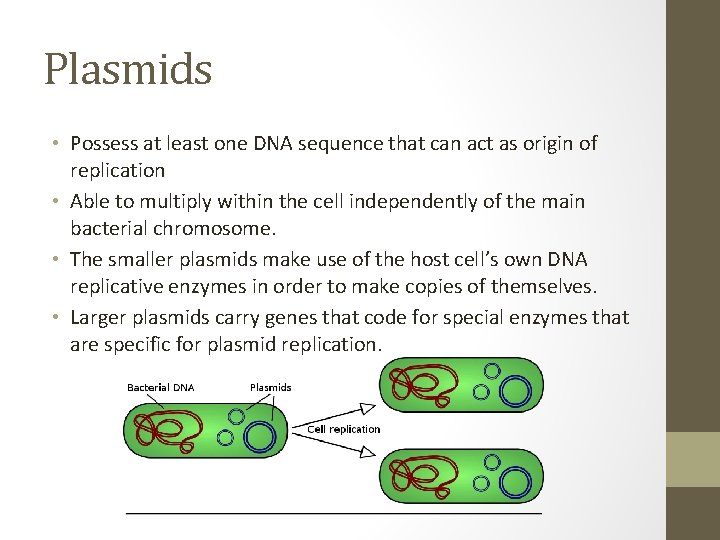
Plasmids • Possess at least one DNA sequence that can act as origin of replication • Able to multiply within the cell independently of the main bacterial chromosome. • The smaller plasmids make use of the host cell’s own DNA replicative enzymes in order to make copies of themselves. • Larger plasmids carry genes that code for special enzymes that are specific for plasmid replication.
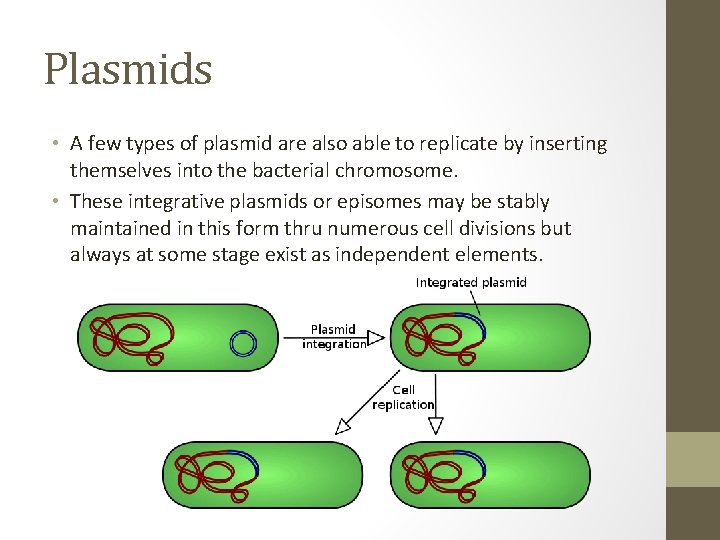
Plasmids • A few types of plasmid are also able to replicate by inserting themselves into the bacterial chromosome. • These integrative plasmids or episomes may be stably maintained in this form thru numerous cell divisions but always at some stage exist as independent elements.

Plasmids: size and copy number • Size and copy number are important. • Less than 10 kb is a desired size for cloning vector. • Plasmids range from about 1 kb (smallest) to 250 kb (largest) are useful for cloning purposes. • Larger plasmids can be adapted for cloning under some circumstances. Plasmid Size (kb) organism p. UC 8 Col. E 1 RP 4 2. 1 6. 4 54 F TOL p. Ti. Ach 5 95 117 213 E. coli Pseudomonas and others E. coli Pseudomonas putida Agrobacterium

Plasmids: size and copy number • Copy number refers to the number of molecules of an individual plasmid that are normally found in a single bacterial cell. • Larger plasmids are stringent- have a low copy number- one or 2 plasmids in a cell. • Others are called relaxed plasmids- present in multiple copies of 50 or more per cell. • A useful cloning vector needs to be present in the cell multiple copies- large quantities of recombinant DNA can be obtained.
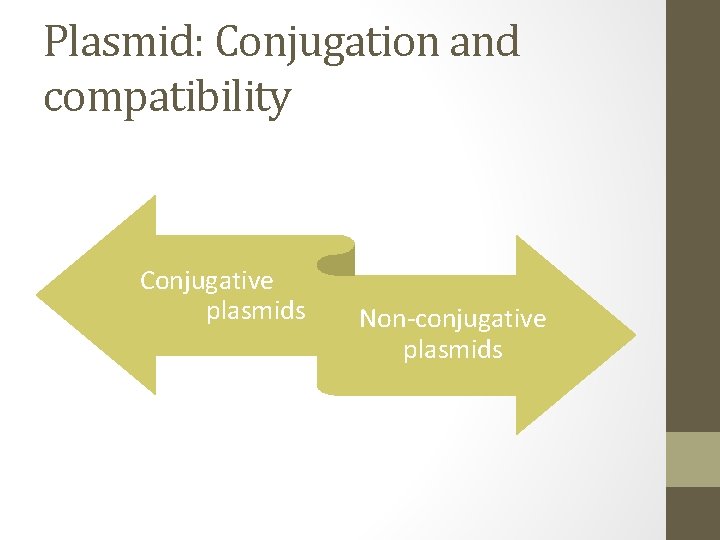
Plasmid: Conjugation and compatibility Conjugative plasmids Non-conjugative plasmids

Conjugative plasmids Conjugative • Ability to promote sexual conjugation between bacterial cells- a process that can result in a conjugative plasmid spreading from one cell to all the other cell in bacterial culture. • Conjugation and plasmid transfer are controlled by a set of genes- tra gene, only present in conjugative plasmids.
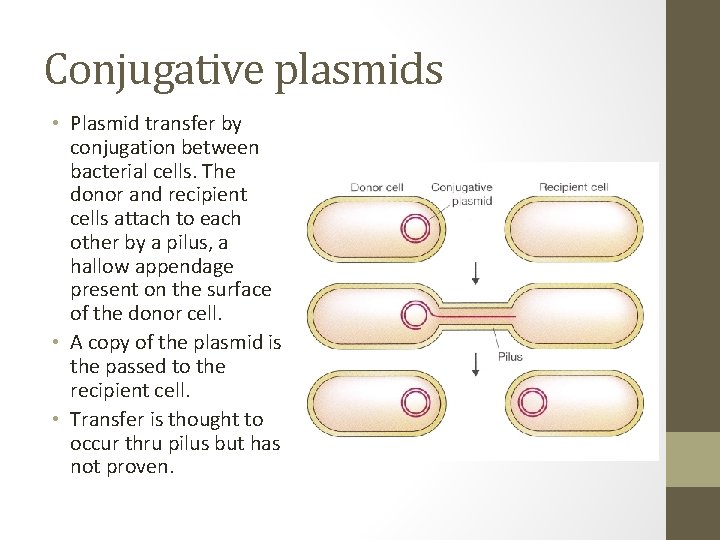
Conjugative plasmids • Plasmid transfer by conjugation between bacterial cells. The donor and recipient cells attach to each other by a pilus, a hallow appendage present on the surface of the donor cell. • A copy of the plasmid is the passed to the recipient cell. • Transfer is thought to occur thru pilus but has not proven.

Plasmid: Conjugation and compatibility • Several different kinds of plasmid may be found in a single cell, including more than 1 conjugative plasmid at 1 time. • E. coli have known to contain 7 different plasmids at once. • To be able to coexist, different plasmids must be compatible. • If 2 plasmids are incompatible, either one will be lost from the cell.

Plasmid classification • The most useful classification: main characteristics coded by the plasmid genes. • The 5 major types of plasmid according to this classification are as follows: 1. Fertility or F plasmids carry only tra genes and have no characteristics beyond the ability to promote conjugal transfer of plasmids. Well known example- F plasmid of E. coli. 2. Resistance or R plasmids- carry genes conferring on the host bacterium resistance to one or more antibacterial agentschloramphenicol, ampicilin and mercury. Example is RP 4 which commonly found in P. putida.
Plasmid classification 3. Col plasmids- code for colicins, protein that kill other bacteria. Example is Col. E 1 of E. coli. 4. Degradative plasmids – allow host bacterium to metabolize unusual molecules such as toluene and salicylic acid. Example TOL of P. putida. 5. Virulence plasmid- confer pathogenicity on host bacterium. Includes Ti plasmid of A. tumefaciens, which induce crown gall disease on dicot plants.
Bacteriophages • Bacteriophages/phages: viruses that specifically infect bacteria. • Phages are : • simple in structure • consisting of merely a DNA (or RNA) molecule carrying a number of genes, including several genes for replication of the phage • Surrounded by a protective coat or capsid made of protein molecules.

2 main types of phage • 2 main types of phage structure • Head-and-tail (e. g: lambda) • Filamentous (e. g. M 13)
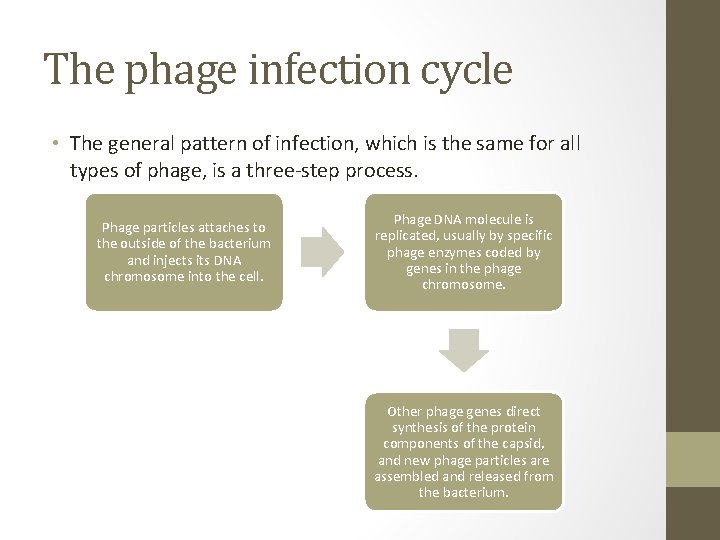
The phage infection cycle • The general pattern of infection, which is the same for all types of phage, is a three-step process. Phage particles attaches to the outside of the bacterium and injects its DNA chromosome into the cell. Phage DNA molecule is replicated, usually by specific phage enzymes coded by genes in the phage chromosome. Other phage genes direct synthesis of the protein components of the capsid, and new phage particles are assembled and released from the bacterium.
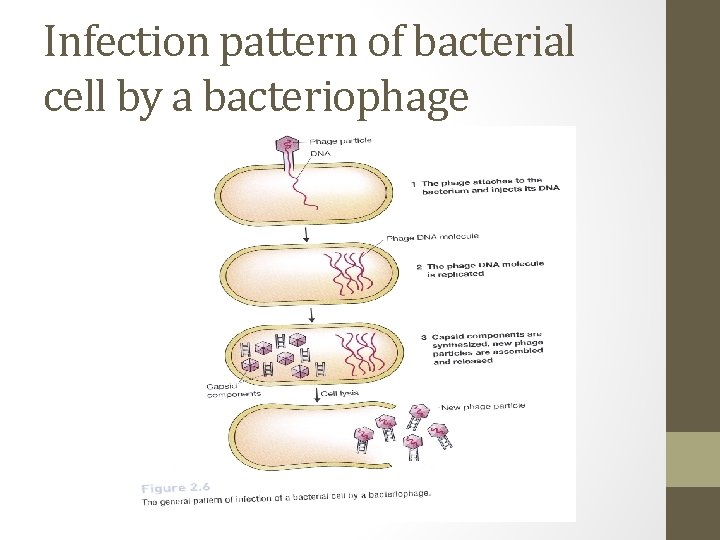
Infection pattern of bacterial cell by a bacteriophage

The phage infection cycle • For some phage, the entire infection cycle is completed very quickly, possibly less than 20 mins. • This rapid infection- lytic cycle, since the release of the new phage particles is associated with lysis of the bacterial cells. • The characteristics feature of a lytic infection cycle is that phage DNA replication is immediately followed by the synthesis of capsid proteins, and phage molecule never maintained in a stable condition in the host cell.
Lysogenic phages • In contrast to lytic cycle, lysogenic infection is characterized by retention of the phage DNA molecule in the host bacterium, possibly for many thousands of cell divisions. • For lysogenic phages: the phage DNA is inserted into the bacterial genome in a manner similar to episomal insertion • The integrated form of the phage DNA- called prophage is usually indistinguishable from an uninfected cell. • However, the prophage is eventually released from the host genome and the phage reverts to the lytic mode and lyses the cell.
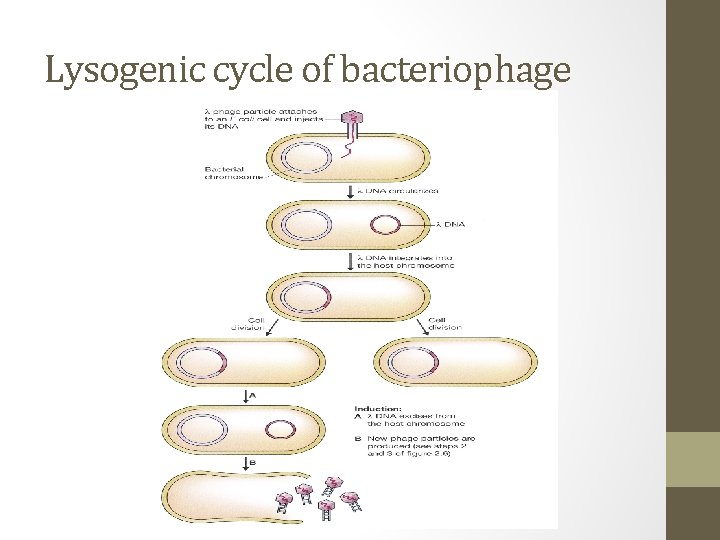
Lysogenic cycle of bacteriophage

Lysogenic phages- M 13 • A limited number of lysogenic phages follow a rather different infection cycle. • M 13 or related phage infects E. coli, new phage particles are continuously assembled and released from the cell. • The M 13 is not integrated into the bacterial genome and does not become quiescent. • With this type of phages, cell lysis never occurs and the infected bacterium can continue to grow and divide but at a slower rate than uninfected cells.
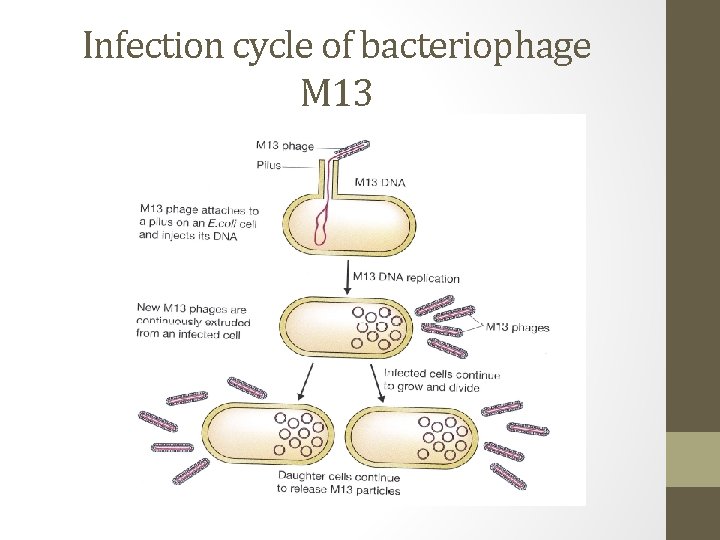
Infection cycle of bacteriophage M 13
Gene organization in the λDNA molecule • λ is a typical example of a head and tail phage. • DNA is contained in the polyhedral head structure- capsid • The tail serves to attach the phage to the bacterial surface and to inject the DNA into the cell. • The genome size of lambda DNA is 49 kb. • The positions and identities of all of the genes in the lambda DNA molecule are known. • A feature of the lambda genetic map: genes related in term of function are clustered together in the genome. • For eg, genes on the left hand of the genome encode for components of the capsid • Middle of the molecule: genes encode for integration of the prophage into host genome.

• Clustering of related genes is important in: - Controlling expression of the lambda genome- it allows genes to be switched on and off as group rather than individually. - Construction of lambda based cloning vectors.
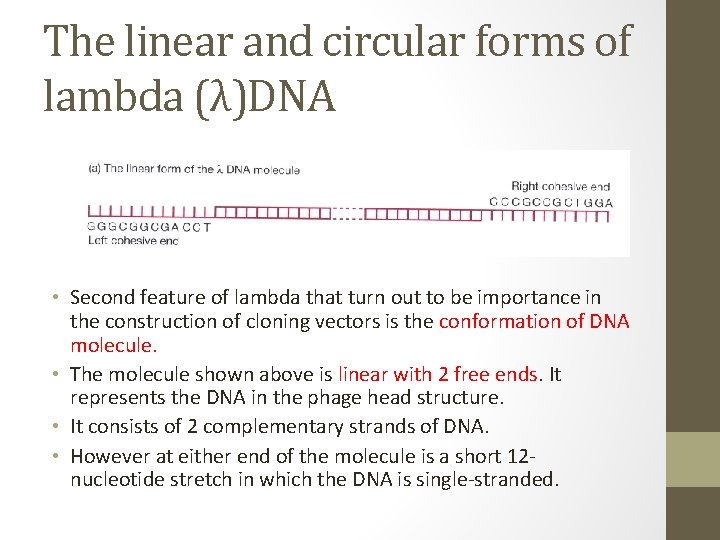
The linear and circular forms of lambda (λ)DNA • Second feature of lambda that turn out to be importance in the construction of cloning vectors is the conformation of DNA molecule. • The molecule shown above is linear with 2 free ends. It represents the DNA in the phage head structure. • It consists of 2 complementary strands of DNA. • However at either end of the molecule is a short 12 nucleotide stretch in which the DNA is single-stranded.
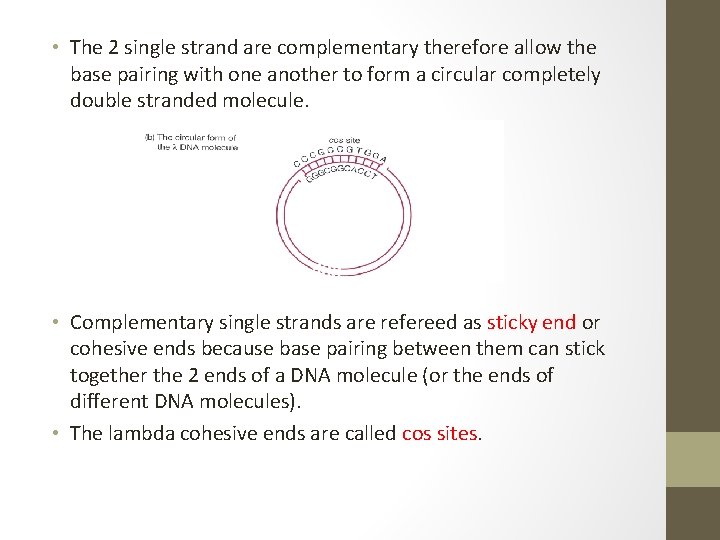
• The 2 single strand are complementary therefore allow the base pairing with one another to form a circular completely double stranded molecule. • Complementary single strands are refereed as sticky end or cohesive ends because base pairing between them can stick together the 2 ends of a DNA molecule (or the ends of different DNA molecules). • The lambda cohesive ends are called cos sites.

Roles of cos sites • Cos sites play 2 distinct roles during lambda infection cycle. 1. They allow the linear DNA molecule that is injected into the cell to be circularized, which is necessary prerequisite for insertion into bacterial genome. 2. After prophage has excised from the host genome, a large number of new lambda DNA molecules are produced by the rolling circle mechanism of replication in which a continuous DNA strand is “rolled off” the template molecule.
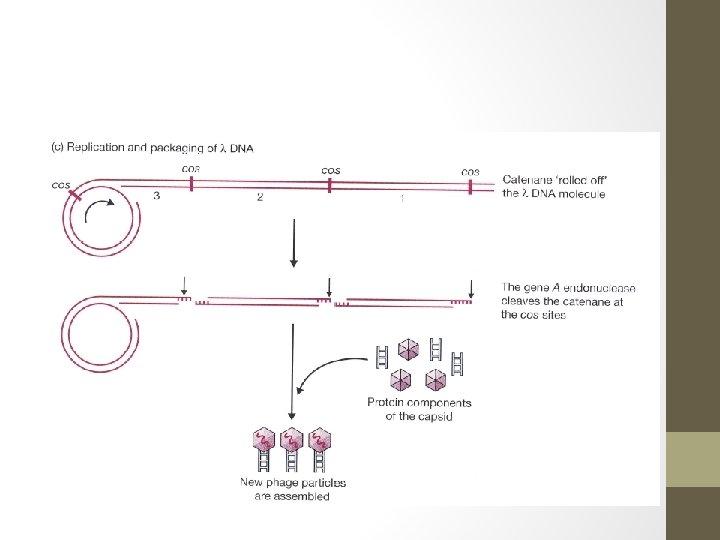

Roles of cos sites - The result is a catenane consisting of a series of linear lambda genomes joined together at the cos sites. - The role of cos sites now is as recognition sequences of endonuclease that cleaves the catenane at the cos sites producing individual lambda genome. - This endonuclease, which is the product of gene A on the lambda DNA molecule creates the single sticky ends and also acts together with other proteins to package lambda genome into a phage head structure. - The cleavage and packaging processes recognize only the cos sites and the DNA sequences to either side of them. - Therefore, changing the structure of the internal regions of the lambda genome has no effect on these events.
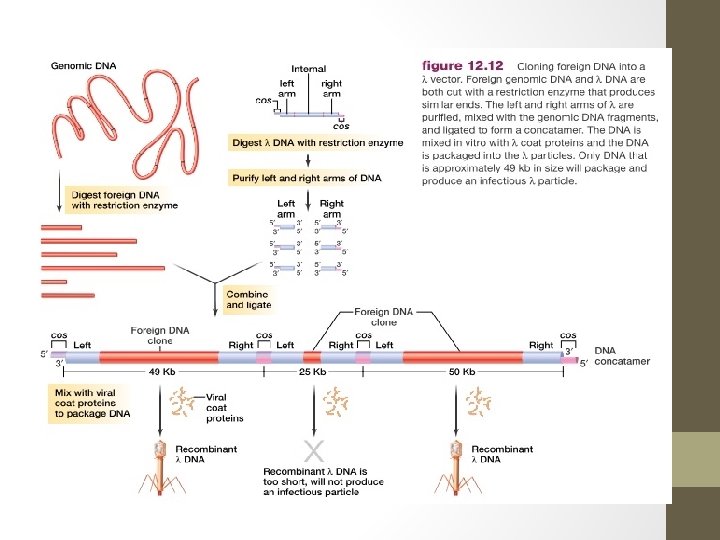
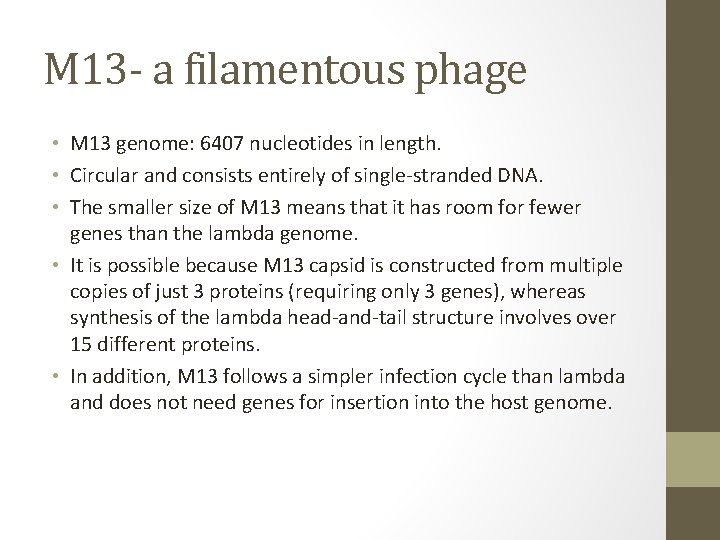
M 13 - a filamentous phage • M 13 genome: 6407 nucleotides in length. • Circular and consists entirely of single-stranded DNA. • The smaller size of M 13 means that it has room for fewer genes than the lambda genome. • It is possible because M 13 capsid is constructed from multiple copies of just 3 proteins (requiring only 3 genes), whereas synthesis of the lambda head-and-tail structure involves over 15 different proteins. • In addition, M 13 follows a simpler infection cycle than lambda and does not need genes for insertion into the host genome.
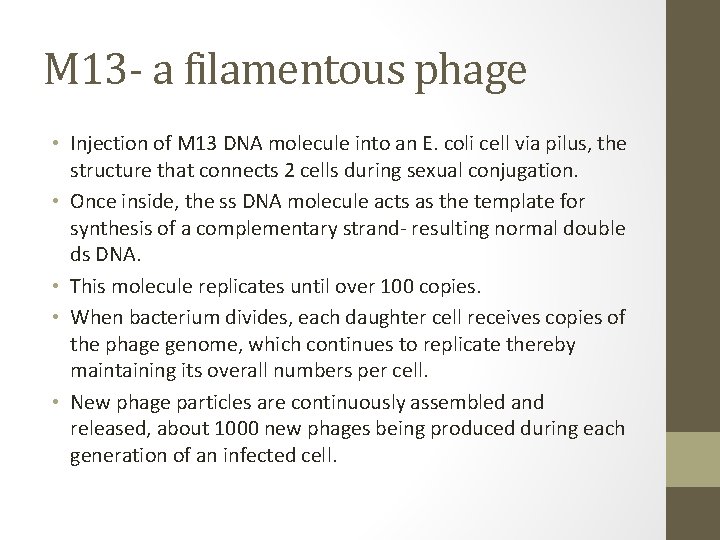
M 13 - a filamentous phage • Injection of M 13 DNA molecule into an E. coli cell via pilus, the structure that connects 2 cells during sexual conjugation. • Once inside, the ss DNA molecule acts as the template for synthesis of a complementary strand- resulting normal double ds DNA. • This molecule replicates until over 100 copies. • When bacterium divides, each daughter cell receives copies of the phage genome, which continues to replicate thereby maintaining its overall numbers per cell. • New phage particles are continuously assembled and released, about 1000 new phages being produced during each generation of an infected cell.
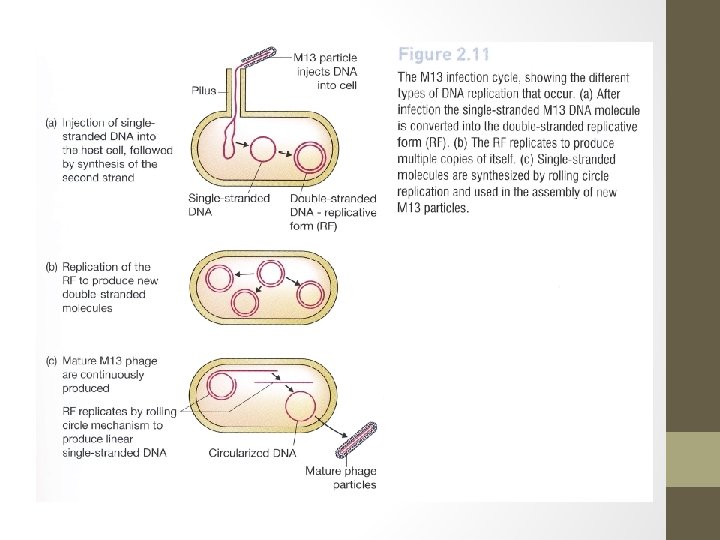
M 13 - a filamentous phage • Several features of M 13 that make this phage attractive as cloning vector. 1. The genome is less than 10 kb in size- within the range desirable for a potential vector. 2. The ds replicative form (RF) of the M 13 genome behaves very much like plasmid, and can be treated as such for experimental purposes. 3. It is easily prepared from a culture of infected E. coli cells and can be reintroduced by transfection. 4. Genes cloned with M 13 -based vector can be obtained in ss form. Ss versions of cloned genes are useful for several techniques ie DNA sequencing & in-vitro mutagenesis. 5. Also used in phage display, a technique for identifying pairs of genes whose protein products interact with one another.

Viruses as cloning vectors for other organisms • Viruses can be used as cloning vector: most living organisms are infected by viruses. • Plasmids use is only limited for bacteria and yeasts but not in higher organisms. • Several eukaryotic viruses have been employed as cloning vectors for specialized applications. • For eg: human adenoviruses are used in gene therapy, baculoviruses used to synthesize important pharmaceutical proteins in insect cells and caulimoviruses and geminiviruses have been used for cloning in plants.
Summary: Practical features of DNA cloning vectors • A DNA molecule needs to display several features to be able to act as cloning vector for gene cloning. • Most of DNA cloning vectors usually include most of the following desirable and practical features: 1. SIZE - Should be small enough to be easily separated from the chromosomal DNA of the host bacteria. 2. Origin of replication (ORI) - The site for DNA replication that allows plasmids to replicate separately from the host cell’s chromosome. - The no of plasmids in the cell- copy number.
Practical features of DNA cloning vectors - Normal copy number of plasmids in most of bacterial cell is small (~12 plasmids/cell). However, many of the desirable plasmids are known as high-copy-number- can replicate to create hundreds/thousands of plasmid copies/cell. 3. Multiple cloning site (MCS) - MCS or polylinker is a stretch of DNA with recognition sequences for many different common restriction enzymes. - These sites are engineered into the plasmid so that digestion of the plasmid with restriction enzymes (RE) does not result in the loss of fragment of DNA. - The circular plasmid becomes linearized when digested with RE.
Practical features of DNA cloning vectors 4. Selectable marker genes - These genes allow for the selection and identification of bacteria that have been transformed with a recombinant plasmid compared to nontransformed cells. - Some of the most common selectable markers are genes for ampicillin resistance (amp. R) and tetracycline resistance (tet. R) and the lac. Z gene used for blue-white screening. 5. RNA polymerase promoter sequences - These sequences are used for transcription of RNA in vivo and in vitro by RNA polymerase. - During transcription, RNA polymerase copies DNA into RNA. In vivo, these sequences allow bacterial cells to make RNA from cloned genes, leads to protein synthesis.

- In vitro, transcribed RNA can be used to synthesize RNA probes that can be used to study gene expression. - 6. DNA sequencing primers sequences - These sequences permit nucleotide sequencing of cloned DNA fragments that have been inserted into the plasmid.
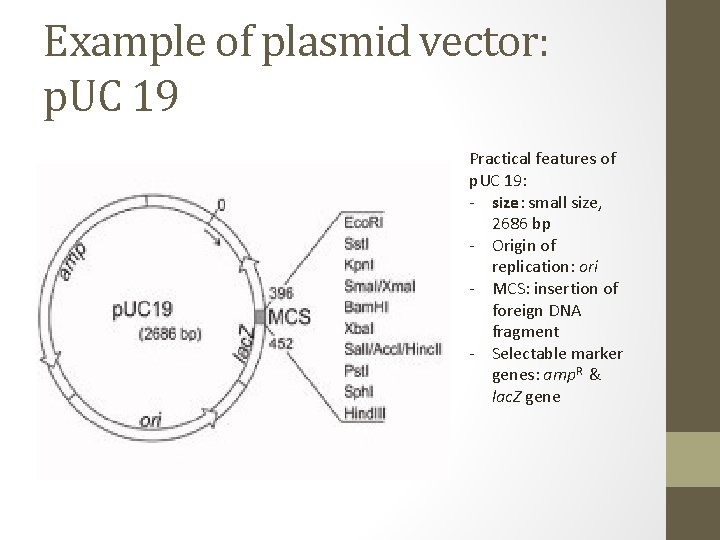
Example of plasmid vector: p. UC 19 Practical features of p. UC 19: - size: small size, 2686 bp - Origin of replication: ori - MCS: insertion of foreign DNA fragment - Selectable marker genes: amp. R & lac. Z gene

- Slides: 42