Using mutants to clone genes Objectives 1 What
Using mutants to clone genes Objectives: 1. What is positional cloning? 2. What is insertional tagging? 3. How can one confirm that the gene cloned is the same one that is mutated to give the phenotype of interest?

Reading • References: • Westhoff et. al. Molecular Plant Development: from gene to plant. Chapter 3: 52 -65.
Positional (map-based) Cloning Map based cloning is a dependable method of cloning a gene using a mutant phenotype, molecular genetic markers and genetic recombination. This method is most easily done in organisms where the necessary tools (genetic map, physical map and or sequence of the genome) are available.

Positional (map-based) Cloning 1. Use the mutant phenotype and DNA-based genetic markers of known position to map, using recombination, the gene of interest to a site on a specific chromosome.
DNA-Based Genetic Markers The genomes of two individuals of the same species are rarely identical and can have many nucleotide differences between them. These variations in DNA sequences often do not alter the function of a gene but can be used as phenotypes in genetic mapping by detecting the differences using: 1. PCR amplification (simple sequence-length polymorphism = SSLP) 2. a combination of both PCR amplification followed by restriction endonuclease digestion (cleaved amplified polymorphic sequences = CAPS). 3. genome sequencing.
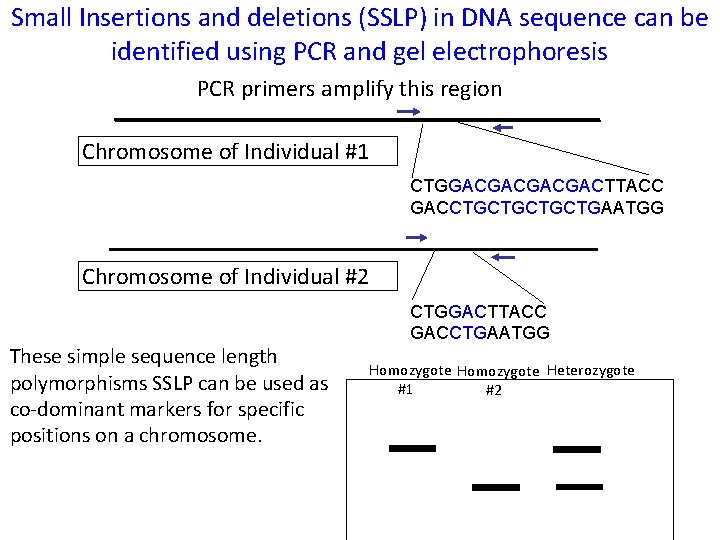
Small Insertions and deletions (SSLP) in DNA sequence can be identified using PCR and gel electrophoresis PCR primers amplify this region Chromosome of Individual #1 CTGGACGACTTACC GACCTGCTGAATGG Chromosome of Individual #2 These simple sequence length polymorphisms SSLP can be used as co-dominant markers for specific positions on a chromosome. CTGGACTTACC GACCTGAATGG Homozygote Heterozygote #1 #2
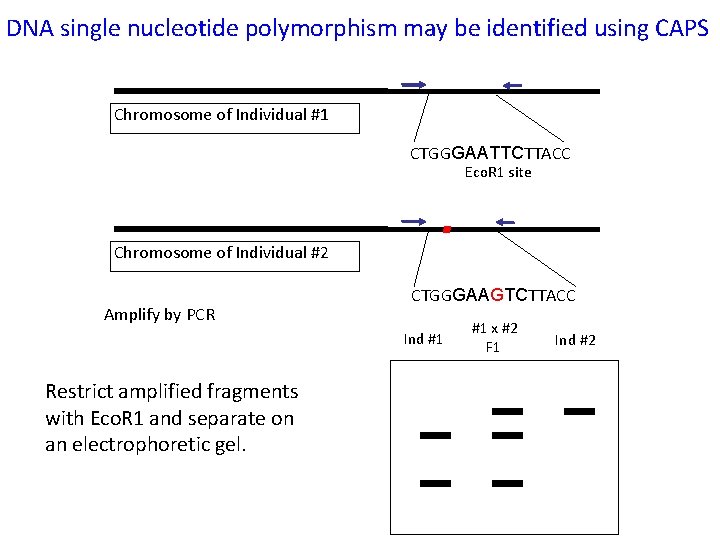
DNA single nucleotide polymorphism may be identified using CAPS Chromosome of Individual #1 CTGGGAATTCTTACC Eco. R 1 site Chromosome of Individual #2 Amplify by PCR CTGGGAAGTCTTACC Ind #1 Restrict amplified fragments with Eco. R 1 and separate on an electrophoretic gel. #1 x #2 F 1 Ind #2
Mapping to DNA-Based Genetic Markers The genomes of individuals form different populations of the same species differ in a large number of SSLPs/CAPS. This variation can be detected and used as genetic markers for specific positions on chromosomes. When two such individuals are crossed all the differences will segregate in the F 2 progeny and can be mapped relative to one another or any novel phenotype in one of the parents. Eg. Arabidopsis populations from different parts of the world are called ecotypes: Columbia (ecotype from southern US)(Col) Landsberg erecta (ecotype from Germany)(Ler)
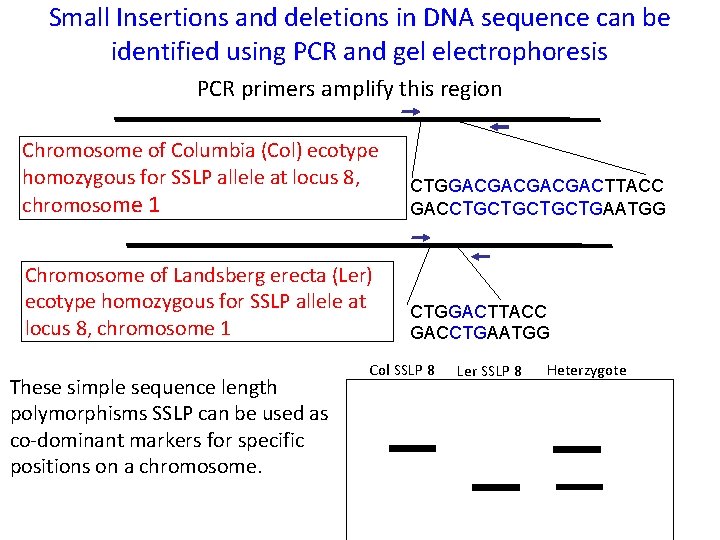
Small Insertions and deletions in DNA sequence can be identified using PCR and gel electrophoresis PCR primers amplify this region Chromosome of Columbia (Col) ecotype homozygous for SSLP allele at locus 8, chromosome 1 CTGGACGACTTACC GACCTGCTGAATGG Chromosome of Landsberg erecta (Ler) ecotype homozygous for SSLP allele at locus 8, chromosome 1 CTGGACTTACC GACCTGAATGG These simple sequence length polymorphisms SSLP can be used as co-dominant markers for specific positions on a chromosome. Col SSLP 8 Ler SSLP 8 Heterzygote

SSLP markers can be mapped using recombination just like genes • In a cross Columbia and Landsberg erecta, the resulting F 1 progeny will be heterozygous at all SSLP loci that were identified between the two: SSLP 16 C/SSLP 16 L; SSLP 71 C/SSLP 71 L; SSLP 8 C/SSLP 8 L Therefore in the F 2 generation they can be mapped relative to one another
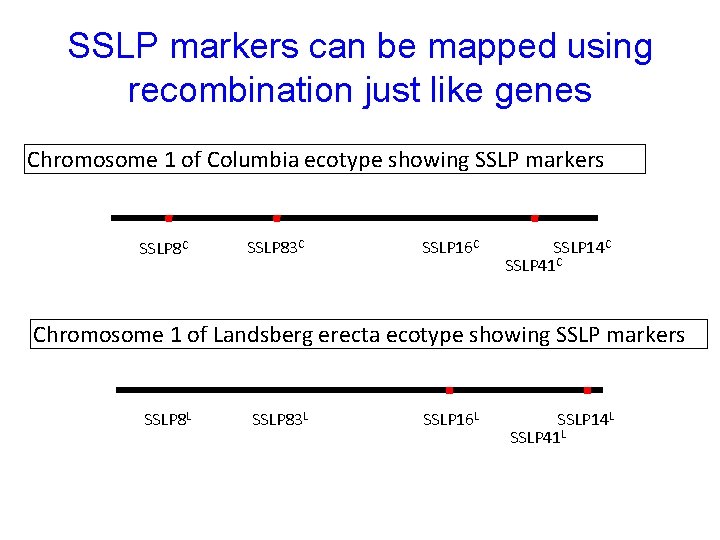
SSLP markers can be mapped using recombination just like genes Chromosome 1 of Columbia ecotype showing SSLP markers SSLP 8 C SSLP 83 C SSLP 16 C SSLP 14 C SSLP 41 C Chromosome 1 of Landsberg erecta ecotype showing SSLP markers SSLP 8 L SSLP 83 L SSLP 16 L SSLP 14 L SSLP 41 L
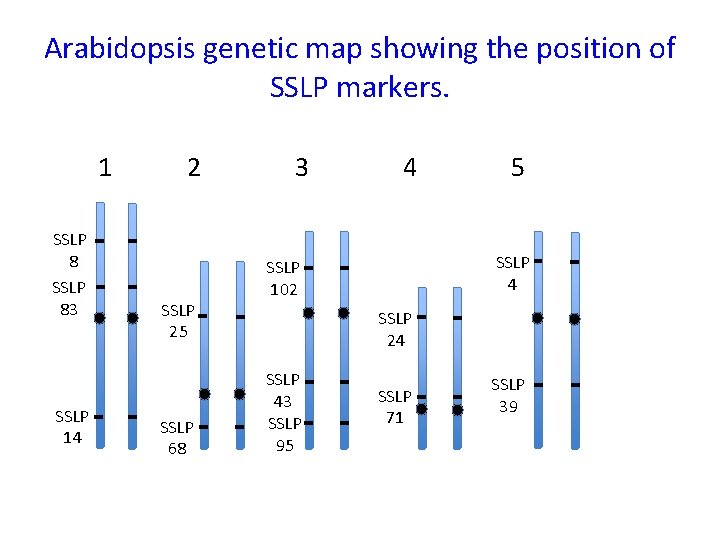
Arabidopsis genetic map showing the position of SSLP markers. 1 SSLP 8 SSLP 83 SSLP 14 2 SSLP 25 SSLP 68 3 4 5 SSLP 4 SSLP 102 SSLP 24 SSLP 43 SSLP 95 SSLP 71 SSLP 39

Apetala 2 mutant has flowers where the sepals and petals are replaced by reproductive organs
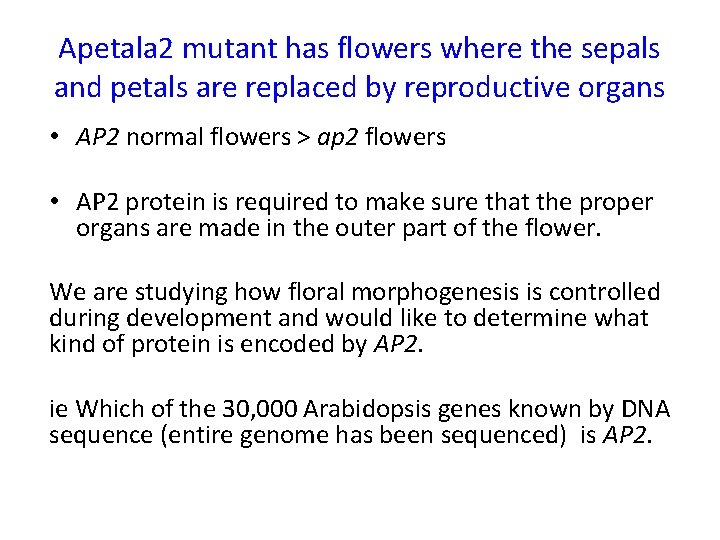
Apetala 2 mutant has flowers where the sepals and petals are replaced by reproductive organs • AP 2 normal flowers > ap 2 flowers • AP 2 protein is required to make sure that the proper organs are made in the outer part of the flower. We are studying how floral morphogenesis is controlled during development and would like to determine what kind of protein is encoded by AP 2. ie Which of the 30, 000 Arabidopsis genes known by DNA sequence (entire genome has been sequenced) is AP 2.
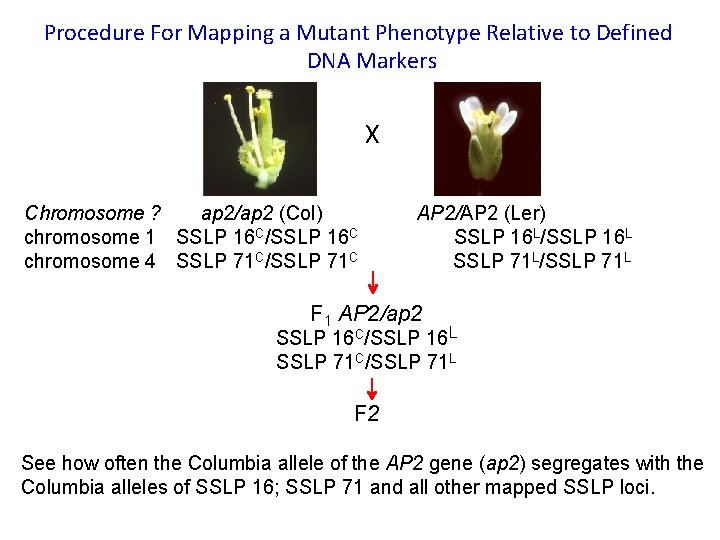
Procedure For Mapping a Mutant Phenotype Relative to Defined DNA Markers X Chromosome ? ap 2/ap 2 (Col) chromosome 1 SSLP 16 C/SSLP 16 C chromosome 4 SSLP 71 C/SSLP 71 C AP 2/AP 2 (Ler) SSLP 16 L/SSLP 16 L SSLP 71 L/SSLP 71 L F 1 AP 2/ap 2 SSLP 16 C/SSLP 16 L SSLP 71 C/SSLP 71 L F 2 See how often the Columbia allele of the AP 2 gene (ap 2) segregates with the Columbia alleles of SSLP 16; SSLP 71 and all other mapped SSLP loci.
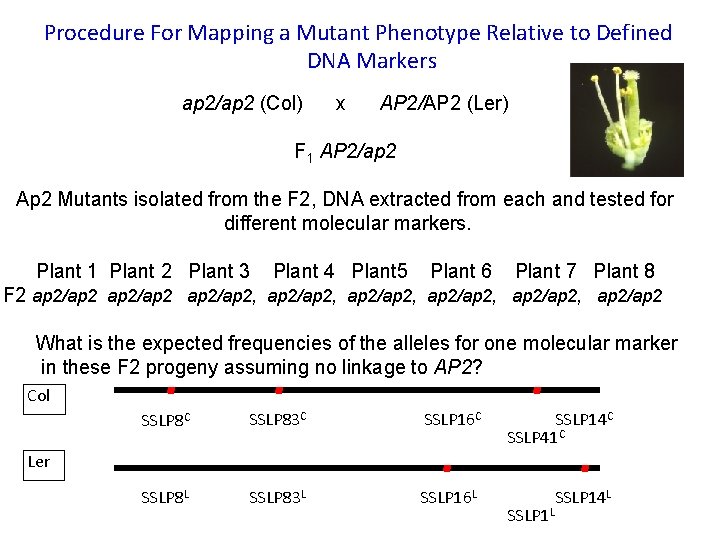
Procedure For Mapping a Mutant Phenotype Relative to Defined DNA Markers ap 2/ap 2 (Col) x AP 2/AP 2 (Ler) F 1 AP 2/ap 2 Ap 2 Mutants isolated from the F 2, DNA extracted from each and tested for different molecular markers. Plant 1 Plant 2 Plant 3 Plant 4 Plant 5 Plant 6 Plant 7 Plant 8 F 2 ap 2/ap 2, ap 2/ap 2, ap 2/ap 2 What is the expected frequencies of the alleles for one molecular marker in these F 2 progeny assuming no linkage to AP 2? Col SSLP 8 C SSLP 83 C SSLP 16 C SSLP 14 C SSLP 41 C SSLP 8 L SSLP 83 L SSLP 16 L SSLP 14 L Ler SSLP 1 L
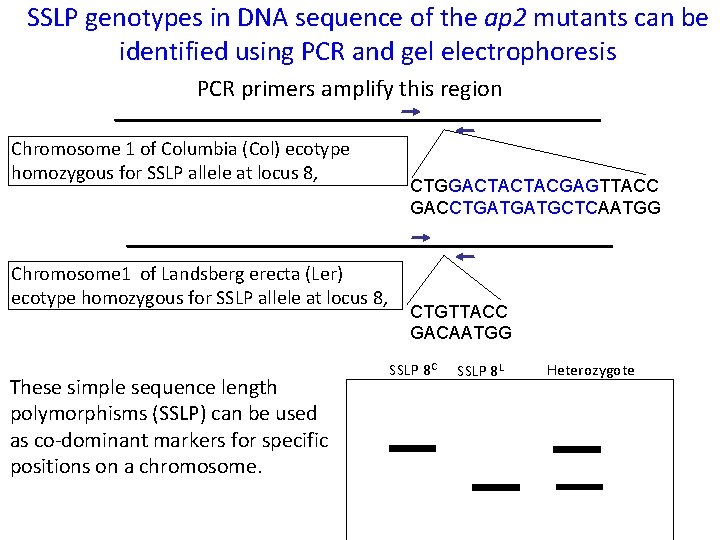
SSLP genotypes in DNA sequence of the ap 2 mutants can be identified using PCR and gel electrophoresis PCR primers amplify this region Chromosome 1 of Columbia (Col) ecotype homozygous for SSLP allele at locus 8, CTGGACTACTACGAGTTACC GACCTGATGATGCTCAATGG Chromosome 1 of Landsberg erecta (Ler) ecotype homozygous for SSLP allele at locus 8, These simple sequence length polymorphisms (SSLP) can be used as co-dominant markers for specific positions on a chromosome. CTGTTACC GACAATGG SSLP 8 C SSLP 8 L Heterozygote
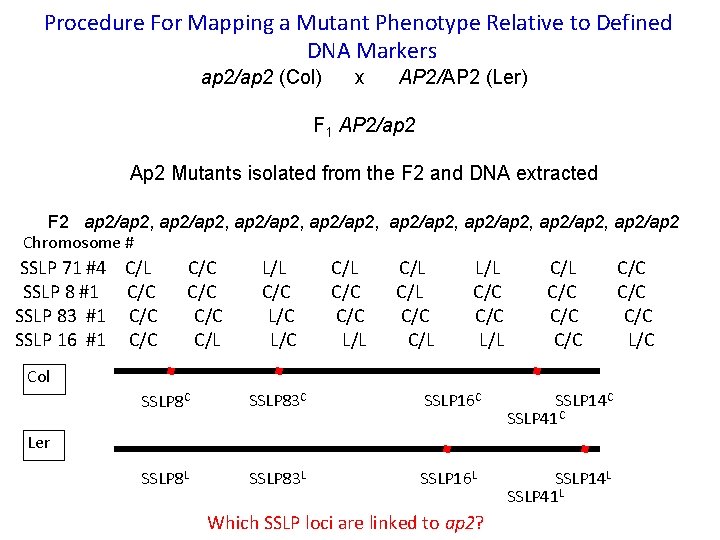
Procedure For Mapping a Mutant Phenotype Relative to Defined DNA Markers ap 2/ap 2 (Col) x AP 2/AP 2 (Ler) F 1 AP 2/ap 2 Ap 2 Mutants isolated from the F 2 and DNA extracted F 2 ap 2/ap 2, ap 2/ap 2, ap 2/ap 2 Chromosome # SSLP 71 #4 C/L C/C L/L C/L L/L C/L C/C SSLP 8 #1 C/C C/L C/C SSLP 83 #1 C/C L/C C/C C/C SSLP 16 #1 C/C C/L L/C L/L C/L L/L C/C L/C Col SSLP 8 C SSLP 83 C SSLP 16 C SSLP 14 C SSLP 41 C SSLP 8 L SSLP 83 L SSLP 16 L SSLP 14 L SSLP 41 L Ler Which SSLP loci are linked to ap 2?

Positional cloning: Pick the best answer**i. Clicker!! Using data from the previous slide, which of the molecular markers is linked to AP 2: A. B. C. D. E. SSLP 71 and SSLP 83 and SSLP 16 SSLP 8 SSLP 71 and SSLP 16
![Chromosome of Columbia ecotype with ap 2 -1 mutation SSLP 8 C [ ] Chromosome of Columbia ecotype with ap 2 -1 mutation SSLP 8 C [ ]](http://slidetodoc.com/presentation_image_h/0893564080e8df17ca5695c15f7eeba0/image-20.jpg)
Chromosome of Columbia ecotype with ap 2 -1 mutation SSLP 8 C [ ] ap 2 -1 SSLP 83 C SSLP 16 C SSLP 14 C SSLP 41 C SSLP 16 L SSLP 14 L SSLP 41 L Chromosome of Landsberg erecta SSLP 8 L AP 2 SSLP 83 L The ap 2 -1 mutant phenotype is found to segregate with SSLP 83 C and SSLP 8 C but no others. The map distance from each of these two to AP 2 is 1/16 = 6. 25 map units. If there is 12. 5 map units between SSLP 83 the AP 2 gene must lie between these two SSLP sites. 1 mu = 270, 000 bp = 60 genes (30, 000 genes in 500 mu) 6. 25 mu = 1, 687, 000 bp with roughly 750 genes.
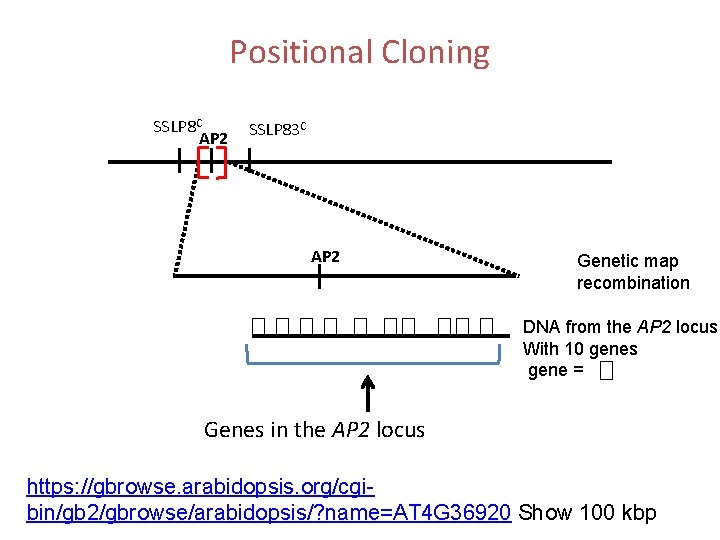
Positional Cloning SSLP 8 C AP 2 SSLP 83 C AP 2 Genetic map recombination DNA from the AP 2 locus With 10 genes gene = Genes in the AP 2 locus https: //gbrowse. arabidopsis. org/cgibin/gb 2/gbrowse/arabidopsis/? name=AT 4 G 36920 Show 100 kbp

The Sequences For All Annotated Genes Are Available • • Sequence: AT 1 G 30960. 1 Date last modified 2003 -05 -27 Name AT 1 G 30960. 1 Tair Accession Sequence: 2015773 Gen. Bank Accession NM_102835 Sequence Length (bp) 1314 Sequence 1 ATGAAAGCTT TTAGATCTCT ACGTATACTA ATTTCCATCT CACGAC 51 GACGAAGACA ACACCTCGTA ATCCCCATCA AGCACAAAAC TTTCTCCGCC 101 GATTTTACTC AGCGCAGCCG AATCTAGACG AACCCACTTC CATCAATGAA 151 GACGGATCAA GCAGCGACTC TGTTTTCGAT AGTAGTCAAT ACCCAATCGA 201 CGATTCCAAT GTAGATTCCG TGAAGAAGCC CAAGGAAGCA ACTTGGGATA 251 AAGGGTACAG AGAAAGAGTA AACAAAGCCT TCTTTGGAAA CTTGACAGAG 301 AAAGGTAAAG TGAAAGTTGC AGAAGAAGAG AGTTCTGAAG ATGATGAGGA 351 TAGTGTTGAT AGGTCAAGGA TTCTCGCTAA GGCTCTCTTA GAGGCTGCGT 401 TAGAGTCACC AGATGAAGAA CTTGGTGAAGTTAG AGAAGAAGAT 451 CAGAAGTCGC TTAATGTCGG CATCATCGGT CCACCTAATG CAGGAAAATC 501 TTCGCTGACT AATTTCATGG TTGGAACAAA GGTTGCTGCT GCTTCACGGA 551 AGACTAACAC GACGACACAT GAAGTGTTAG GAGTATTGAC AAAAGGAGAT 601 ACACAAGTCT GTTTCTTCGA TACTCCGGGT CTGATGCTGA AGAAAAGCGG 651 ATATGGTTAC AAAGACATCA AGGCTCGTGT GCAAAATGCT TGGACTTCTG 701 TTGACCTGTT TGATGTCCTC ATTGTTATGT TTGATGTCCA TAGGCATCTC 751 ATGAGTCCCG ATTCAAGAGT GGTACGCTTG ATCAAATACA TGGGAGAAGA 801 AGAAAATCCG AAACAAAAGC GCGTTTTATGAACAAA GTTGATCTGG 851 TTGAGAAGAA AAAGGATCTA TTAAAGGTTG CTGAGGAGTT CCAAGATCTT 901 CCGGCATATG AAAGATACTT CATGATATCG GGACTTAAGG GATCAGGAGT 951 GAAAGATCTT TCCCAATACT TAATGGATCA GGCTGTTAAA AAACCATGGG 1001 AAGAAGATGC ATTCACGATG AGTGAAGAAG TCTTGAAGAA CATTTCTCTT 1051 GAAGTTGTTA GGGAGAGATT ACTAGACCAT GTCCATCAGG AAATACCATA 1101 TGGTCTGGAG CACCGTCTAG TGGACTGGAA AGAGCTGCGT GACGGGTCTC 1151 TTAGAATTGA ACAGCATCTC ATCACTCCTA AACTTAGCCA ACGCAAGATT 1201 CTTGTAGGCA AGGGCGGTTG CAAGATCGGG AGGATAGGAA TTGAGGCCAA 1251 TGAAGAACTC AGGAGAATAA TGAACCGCAA AGTTCATCTC ATTCTCCAGG 1301 TTAAGCTCAA GTGA Comments (shows only the most recent comments by default) Attribution type name datesubmitted_by AGI-TIGR 2001 -03 -06 submitted_by Gen. Bank 2002 -08 -20 General comments or questions: curator@arabidopsis. org Seed or DNA stock questions (donations, availability, orders, etc): abrc@arabidopsis. org
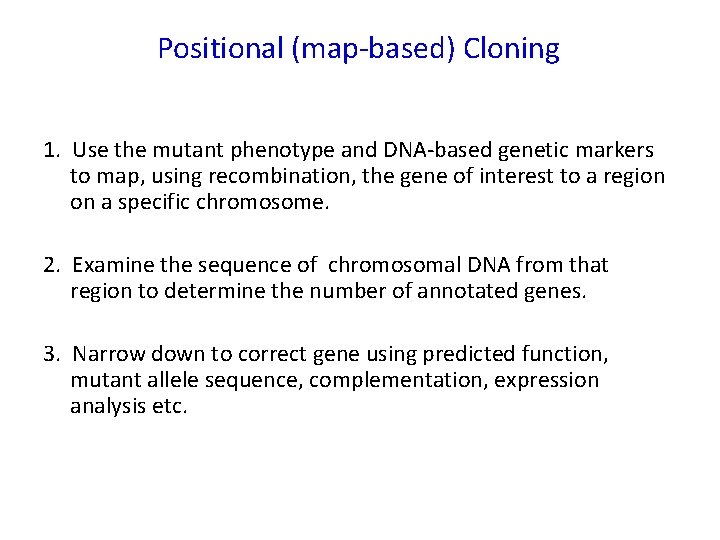
Positional (map-based) Cloning 1. Use the mutant phenotype and DNA-based genetic markers to map, using recombination, the gene of interest to a region on a specific chromosome. 2. Examine the sequence of chromosomal DNA from that region to determine the number of annotated genes. 3. Narrow down to correct gene using predicted function, mutant allele sequence, complementation, expression analysis etc.
Insertional Tagging 1. Isolate mutant phenotype of interest from an insertional mutagenized population of plants. (Insertion DNA must be cloned: eg TDNA or Transposon). 2. Check that the transposon or TDNA in the mutant co-segregates with the mutant phenotype. ---The segregation of an insert can often be followed using the phenotype of a gene encoded in the insert (eg Kanamycin resistance-remember this is a dominant marker), a probe for the insert or PCR primers that can amplify part of the insert. ---repetitive elements (eg. transposons) may complicate such an analysis.
Insertional Tagging 1. Isolate mutant phenotype of interest from an insertional mutagenized population of plants. (Insertion DNA must be cloned: eg TDNA or Transposon). 2. Check that the transposon or TDNA in the mutant segregates with the mutant phenotype. 3. Clone or amplify the chromosomal DNA at the site of insertion using the known sequence of the TDNA or transposon.
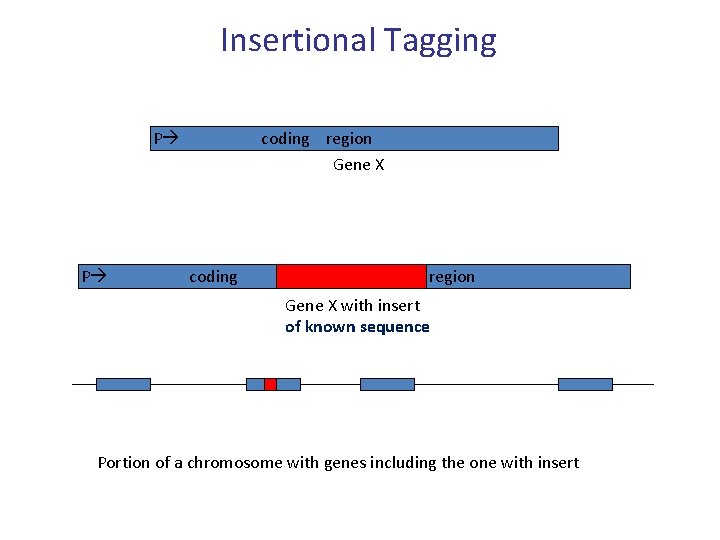
Insertional Tagging P coding region Gene X P coding region Gene X with insert of known sequence Portion of a chromosome with genes including the one with insert
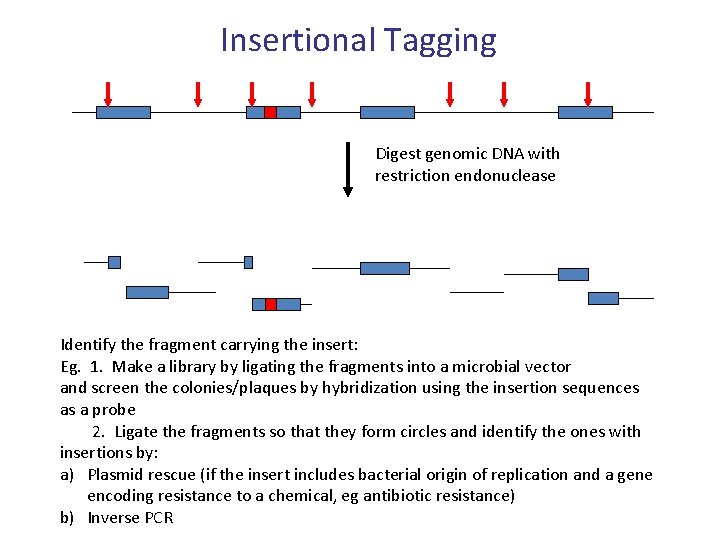
Insertional Tagging Digest genomic DNA with restriction endonuclease Identify the fragment carrying the insert: Eg. 1. Make a library by ligating the fragments into a microbial vector and screen the colonies/plaques by hybridization using the insertion sequences as a probe 2. Ligate the fragments so that they form circles and identify the ones with insertions by: a) Plasmid rescue (if the insert includes bacterial origin of replication and a gene encoding resistance to a chemical, eg antibiotic resistance) b) Inverse PCR
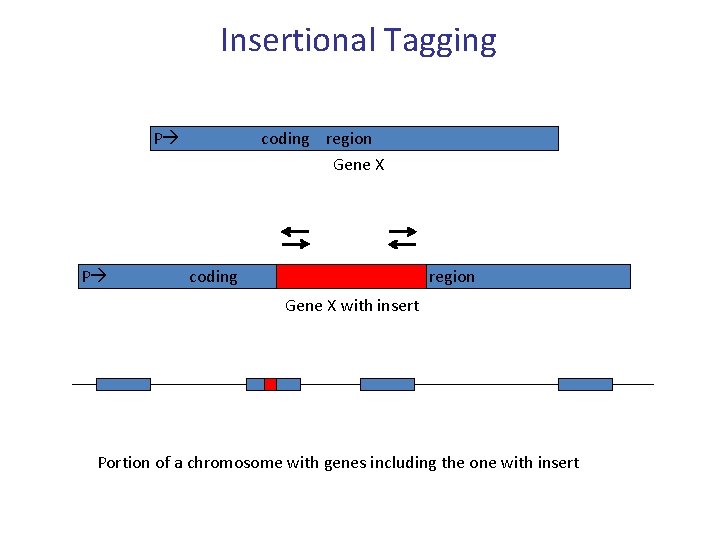
Insertional Tagging P coding region Gene X P coding region Gene X with insert Portion of a chromosome with genes including the one with insert
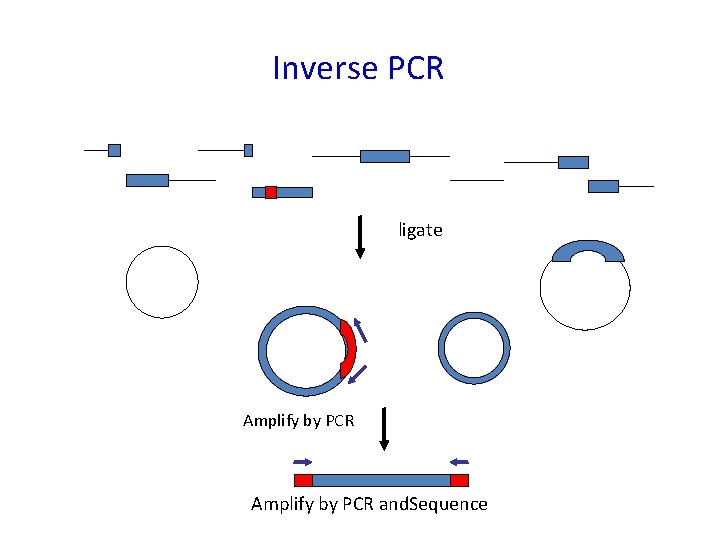
Inverse PCR ligate Amplify by PCR and. Sequence
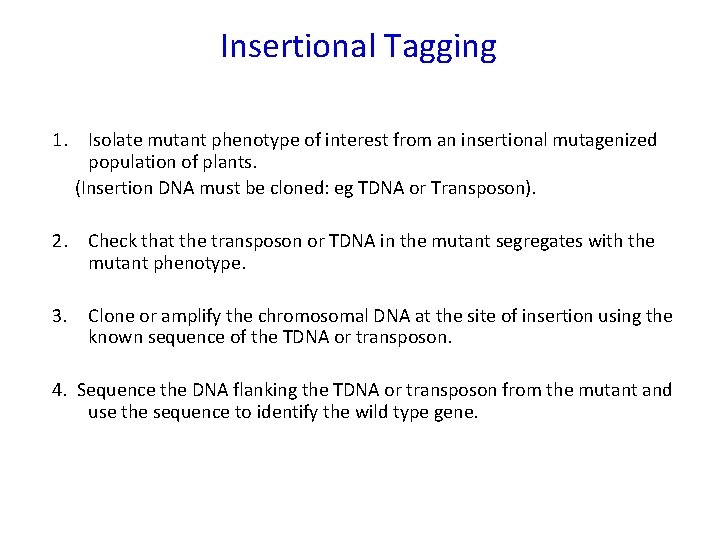
Insertional Tagging 1. Isolate mutant phenotype of interest from an insertional mutagenized population of plants. (Insertion DNA must be cloned: eg TDNA or Transposon). 2. Check that the transposon or TDNA in the mutant segregates with the mutant phenotype. 3. Clone or amplify the chromosomal DNA at the site of insertion using the known sequence of the TDNA or transposon. 4. Sequence the DNA flanking the TDNA or transposon from the mutant and use the sequence to identify the wild type gene.
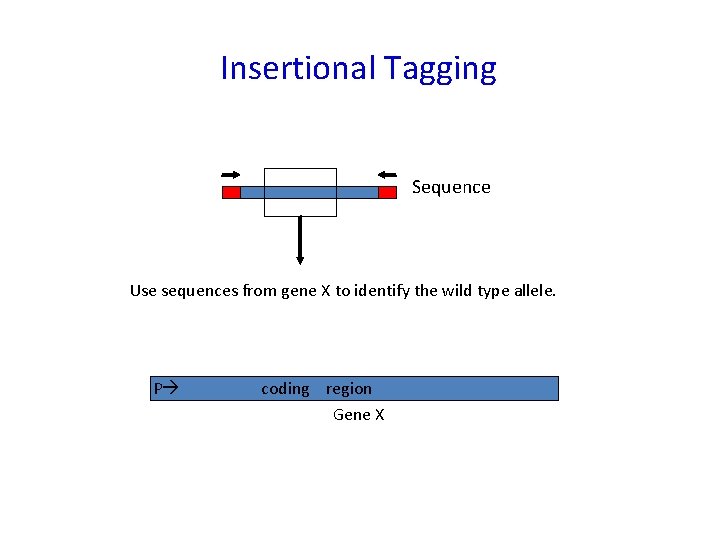
Insertional Tagging Sequence Use sequences from gene X to identify the wild type allele. P coding region Gene X
Connecting a cloned gene with a mutant phenotype Despite the method of identifying the DNA, one must confirm that the gene DNA sequence (X) is the same gene that is mutated to give the phenotype of interest (gene M). Approaches for verifying that SBEI was really the ‘R’ gene were used in the Bhattacharyya et al, 1990 paper we will do in tutorial? What were they?
Connecting a cloned gene with a mutant phenotype Despite the method of identifying the DNA, one must confirm that the gene DNA sequence (X) is the same gene that is mutated to give the phenotype of interest (gene M). 1. Trans gene complementation 2. Sequencing gene derived from each mutant 3. Co-segregation analysis between gene sequence and the mutant phenotype 4. Reverse genetics

Connecting a cloned gene with a mutant phenotype Despite the method of identifying the DNA, one must confirm that the gene DNA sequence (X) is the same gene that is mutated to give the phenotype of interest (gene M). 1. Trans gene complementation Transform a wild type copy of gene X into the mutant and determine if the transgene can rescue (complement) the mutant phenotype (if the mutant allele is recessive) OR transform the mutant allele of gene X into the wild type to see if the transgene will result in the mutant phenotype (if the mutant allele is dominant to wild type) Eg. If we believe that the AP 2 gene (identified by floral phenotrype) encodes the MYB 83 transcription factor we can transform the ap 2 mutant (ap 2 is recessive to AP 2) with a DNA fragment carrying the wild type allele of MYB 83 gene. The transformed plant should have a wild type phenotype if MYB 83 is really the AP 2 gene.
Connecting a cloned gene with a mutant phenotype 1. Transgene complementation. 2. Sequence gene X from several mutants homozygous for different alleles of gene M (if multiple alleles exist). If the ‘M’ gene and X gene are the same then one should find a gene X mutation in every M mutant. This hypothesis can be tested by sequencing gene X from the M mutant. If you have several mutants each of which is homozygous for a different allele then you should find that each mutant has a different mutation in gene X.
Connecting a cloned gene with a mutant phenotype Example: Genotype of plants homozygous for different alleles of the AP 2 gene: • AP 2/AP 2 ap 2 -1/ap 2 -1 ap 2 -2/ap 2 -2 -cloned a wild type gene, MYB 83, encoding a transcription factor. Is MYB 83 gene AP 2? Amplify MYB 83 from plants with each of the genotypes above by PCR amplification. If MYB 83 is AP 2 then • MYB 83 from AP 2/AP 2 will have a wild type sequence. • MYB 83 from ap 2 -1/ap 2 -1 will have a mutation. • MYB 83 from ap 2 -2/ap 2 -2 will also have a mutation but different from that of ap 2 -1.
Connecting a cloned gene with a mutant phenotype 1. Transgene complementation. 2. Sequence gene X from several mutants homozygous for different alleles of gene M. 3. Cosegregation analysis. DNA-based markers (RFLP) identifying gene X should cosegregate with the mutant phenotype M in genetic crosses. This hypothesis can be tested by crossing mutant M to a wild type plant, self-fertilizing the F 1 progeny to produce F 2 progeny and scoring F 2 plants for the mutant phenotype and the gene X molecular marker. Eg. Follow the segregation of a molecular marker for the MYB 83 gene with the ap 2 mutant phenotype. Ordinarily one would not expect to find any recombinants between alleles of AP 2 and MYB 83.
Connecting a cloned gene with a mutant phenotype 1. Transgene complementation. 2. Sequence gene X from several mutants homozygous for different alleles of gene M. 3. Cosegregation analysis. 4. Reverse genetics. Identify mutant alleles of gene X using reverse genetics. Mutations in gene X should have the same phenotype as mutant M and fail to complement the M mutant phenotype. Eg. A loss of function mutation in the MYB 83 gene should have an ap 2 mutant phenotype.
- Slides: 38