Using HDF 5 and Python The H 5

![Python has lists: >>> for elem in ['First. Item', 'Second. Item', 'Third. Item']: . Python has lists: >>> for elem in ['First. Item', 'Second. Item', 'Third. Item']: .](https://slidetodoc.com/presentation_image/d4b89b19b2f24b7382c96868ecd4fb42/image-2.jpg)











- Slides: 23

Using HDF 5 and Python: The H 5 py module Daniel Kahn Science Systems and Applications, Inc. Acknowledgement: Thanks to Ed Masuoka, NASA Contract NNG 06 HX 18 C HDF & HDF-EOS Workshop XV 17 April 2012
![Python has lists for elem in First Item Second Item Third Item Python has lists: >>> for elem in ['First. Item', 'Second. Item', 'Third. Item']: .](https://slidetodoc.com/presentation_image/d4b89b19b2f24b7382c96868ecd4fb42/image-2.jpg)
Python has lists: >>> for elem in ['First. Item', 'Second. Item', 'Third. Item']: . . . print elem. . . First. Item Second. Item Third. Item >>> We can assign the list to a variable. >>> My. List = ['First. Item', 'Second. Item', 'Third. Item'] >>> for elem in My. List: . . . print elem. . . First. Item Second. Item Third. Item HDF & HDF-EOS Workshop XV 17 April 2012 >>>

Lists can contain a mix of objects: >>> Mixed. List = ['My. String', 5, [72, 99. 44]] >>> for elem in Mixed. List: . . . print elem. . . My. String A list inside a list 5 [72, 99. 44] Lists can be addressed by index: >>> Mixed. List[0] 'My. String' >>> Mixed. List[2] [72, 99. 44] HDF & HDF-EOS Workshop XV 17 April 2012

A note about Python lists: Python lists are one dimensional. Arithmetic operations don’t work on them. Don’t be tempted to use them for scientific array based data sets. More the ‘right way’ later. . . HDF & HDF-EOS Workshop XV 17 April 2012

Python has dictionaries. Dictionaries are key, value pairs >>> Dictionary = {'First. Key': 'First. Value', 'Second. Key': 'Second. Value', 'Third. Key': 'Third. Value'} >>> Dictionary {'Second. Key': 'Second. Value', 'Third. Key': 'Third. Value', 'First. Key': 'First. Value'} >>> Notice that Python prints the key, value pairs in a different order than I typed them. The Key, Value pairs in a dictionary are unordered. HDF & HDF-EOS Workshop XV 17 April 2012

Dictionaries are not lists, however we can easily create a list of the dictionary keys: >>> list(Dictionary) ['Second. Key', 'Third. Key', 'First. Key'] >>> We can use a dictionary in a loop without additional elaboration: >>> for Key in Dictionary: . . . print Key, "---->", Dictionary[Key]. . . Second. Key ----> Second. Value Third. Key ----> Third. Value First. Key ----> First. Value >>> HDF & HDF-EOS Workshop XV 17 April 2012

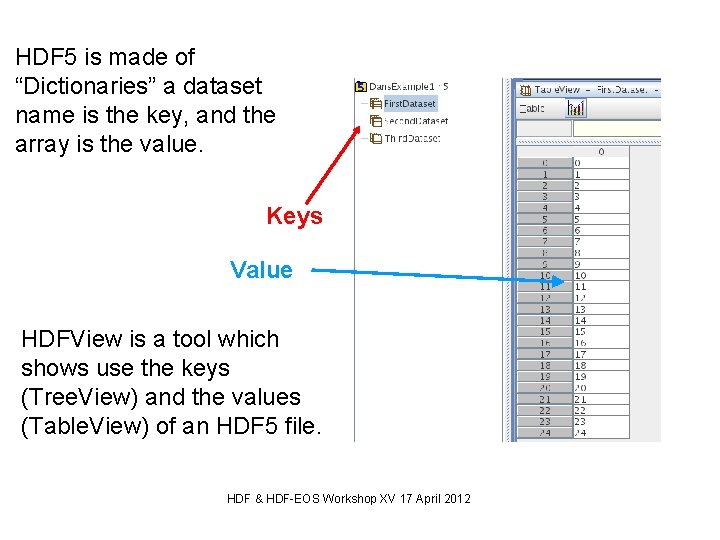
HDF 5 is made of “Dictionaries” a dataset name is the key, and the array is the value. Keys Value HDFView is a tool which shows use the keys (Tree. View) and the values (Table. View) of an HDF 5 file. HDF & HDF-EOS Workshop XV 17 April 2012

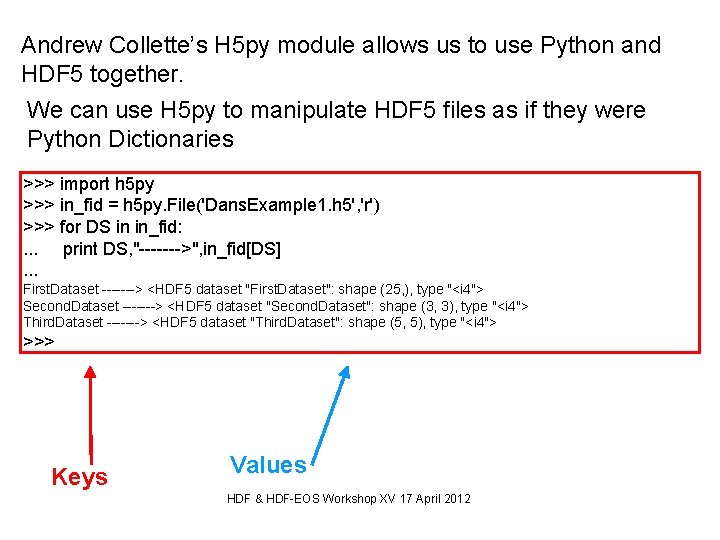
Andrew Collette’s H 5 py module allows us to use Python and HDF 5 together. We can use H 5 py to manipulate HDF 5 files as if they were Python Dictionaries >>> import h 5 py >>> in_fid = h 5 py. File('Dans. Example 1. h 5', 'r') >>> for DS in in_fid: . . . print DS, "------->", in_fid[DS]. . . First. Dataset -------> <HDF 5 dataset "First. Dataset": shape (25, ), type "<i 4"> Second. Dataset -------> <HDF 5 dataset "Second. Dataset": shape (3, 3), type "<i 4"> Third. Dataset -------> <HDF 5 dataset "Third. Dataset": shape (5, 5), type "<i 4"> >>> Keys Values HDF & HDF-EOS Workshop XV 17 April 2012

So What? We need to be able to manipulate the arrays, not just the file. The Numpy module by Travis Oliphant allows the manipulation of arrays in Python. We will see examples of writing arrays later, but to get arrays from the H 5 py object we have the ellipses. >>> import h 5 py >>> fid = h 5 py. File('Dans. Example 1. h 5', 'r') >>> fid['First. Dataset'] <HDF 5 dataset "First. Dataset": shape (25, ), type "<i 4"> >>> fid['First. Dataset'][. . . ] array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24]) >>> type(fid['First. Dataset'][. . . ]) <type 'numpy. ndarray'> >>> HDF & HDF-EOS Workshop XV 17 April 2012

Reasons to use Python and HDF 5 instead of C or Fortran The basic Python Dictionary object has a close similarity to the HDF 5 Group. The object oriented and dynamic nature of Python allows the existing Dictionary syntax to be repurposed for HDF 5 manipulation. In short, working with HDF 5 in Python requires much less code than C or Fortran which means faster development and fewer errors. HDF & HDF-EOS Workshop XV 17 April 2012

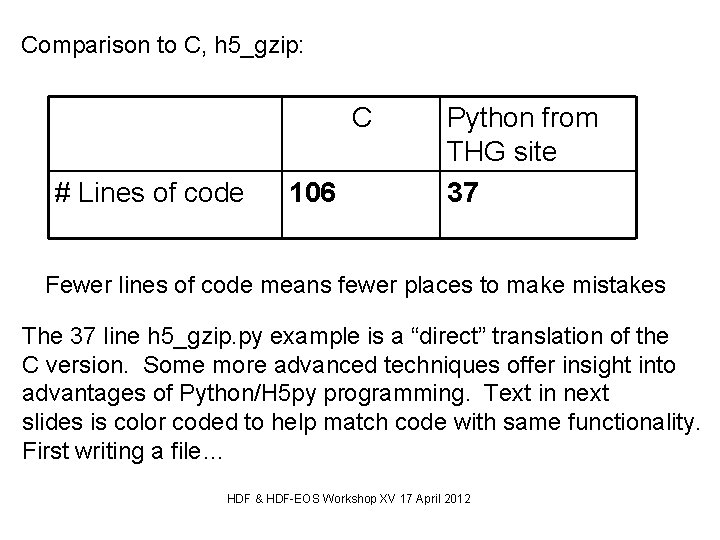
Comparison to C, h 5_gzip: C # Lines of code 106 Python from THG site 37 Fewer lines of code means fewer places to make mistakes The 37 line h 5_gzip. py example is a “direct” translation of the C version. Some more advanced techniques offer insight into advantages of Python/H 5 py programming. Text in next slides is color coded to help match code with same functionality. First writing a file… HDF & HDF-EOS Workshop XV 17 April 2012

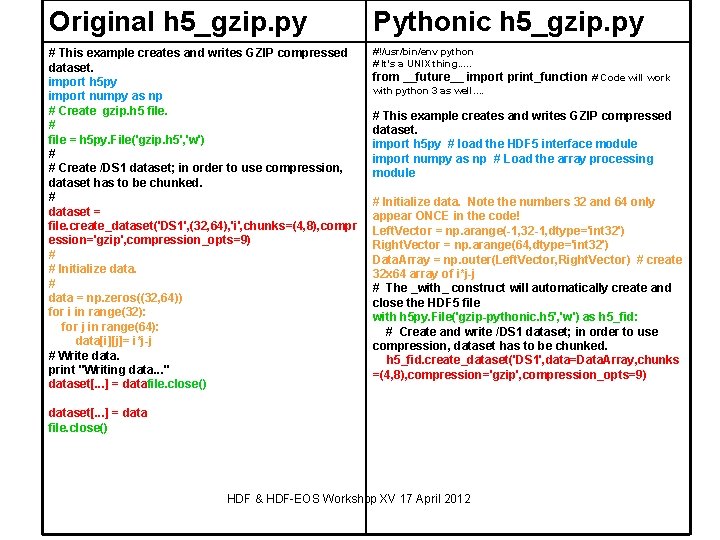
Original h 5_gzip. py Pythonic h 5_gzip. py # This example creates and writes GZIP compressed dataset. import h 5 py import numpy as np # Create gzip. h 5 file. # file = h 5 py. File('gzip. h 5', 'w') # # Create /DS 1 dataset; in order to use compression, dataset has to be chunked. # dataset = file. create_dataset('DS 1', (32, 64), 'i', chunks=(4, 8), compr ession='gzip', compression_opts=9) # # Initialize data. # data = np. zeros((32, 64)) for i in range(32): for j in range(64): data[i][j]= i*j-j # Write data. print "Writing data. . . " dataset[. . . ] = datafile. close() #!/usr/bin/env python # It's a UNIX thing. . . from __future__ import print_function # Code will work with python 3 as well. . # This example creates and writes GZIP compressed dataset. import h 5 py # load the HDF 5 interface module import numpy as np # Load the array processing module # Initialize data. Note the numbers 32 and 64 only appear ONCE in the code! Left. Vector = np. arange(-1, 32 -1, dtype='int 32') Right. Vector = np. arange(64, dtype='int 32') Data. Array = np. outer(Left. Vector, Right. Vector) # create 32 x 64 array of i*j-j # The _with_ construct will automatically create and close the HDF 5 file with h 5 py. File('gzip-pythonic. h 5', 'w') as h 5_fid: # Create and write /DS 1 dataset; in order to use compression, dataset has to be chunked. h 5_fid. create_dataset('DS 1', data=Data. Array, chunks =(4, 8), compression='gzip', compression_opts=9) dataset[. . . ] = data file. close() HDF & HDF-EOS Workshop XV 17 April 2012

Reading data…. # Read data back; display compression properties and dataset max value. # file = h 5 py. File('gzip. h 5', 'r') dataset = file['DS 1'] print "Compression method is", dataset. compression print "Compression parameter is", dataset. compression_opts data = dataset[. . . ] print "Maximum value in", dataset. name, "is: ", max(data. ravel()) file. close() # Read data back; display compression properties and dataset max value. # with h 5 py. File('gzip-pythonic. h 5', 'r') as h 5_fid: dataset = h 5_fid['DS 1'] print("Compression method is", dataset. compression) print("Compression parameter is", dataset. compression_opts) print("Maximum value in", dataset. name, "is: ", dataset. value. max()) HDF & HDF-EOS Workshop XV 17 April 2012

And finally, just to see what the file looks like… HDF & HDF-EOS Workshop XV 17 April 2012

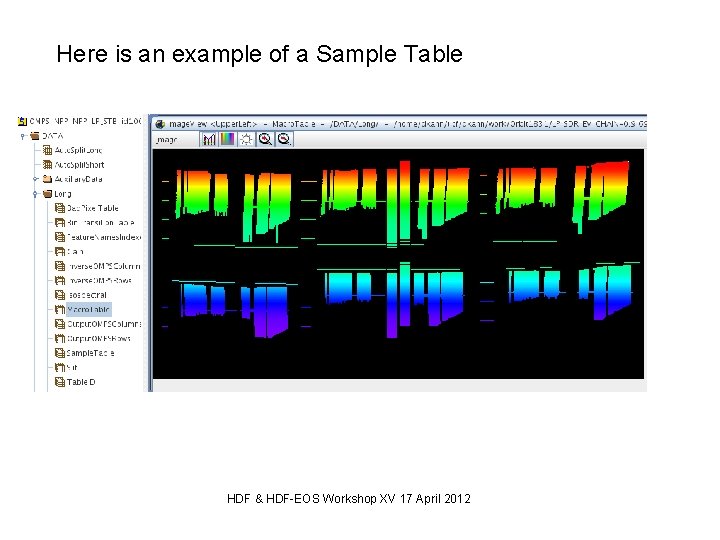
Real world example: Table Comparison Background: For the OMPS Instruments we need to design binary arrays to be uploaded to the satellite to sub-sample the CCD to reduced data rate. For ground processing use we store these arrays in HDF 5. As part of the design process we want to be able to compare arrays in two different files. HDF & HDF-EOS Workshop XV 17 April 2012

Here is an example of a Sample Table HDF & HDF-EOS Workshop XV 17 April 2012

Here is another example: HDF & HDF-EOS Workshop XV 17 April 2012

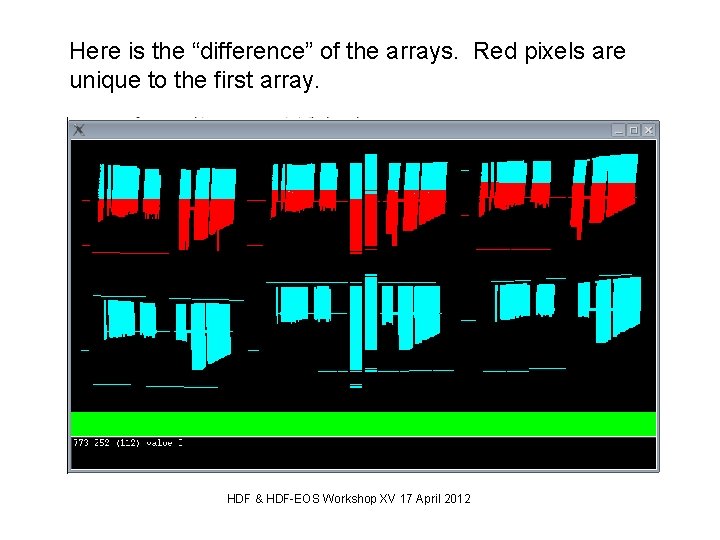
Here is the “difference” of the arrays. Red pixels are unique to the first array. HDF & HDF-EOS Workshop XV 17 April 2012

The code: Compare. ST. py #!/usr/bin/env python """ Documentation """ from __future__ import print_function, division import h 5 py import numpy import View. Frame def Compare. ST(ST 1, ST 2, Int. Time): with h 5 py. File(ST 1, 'r') as st 1_fid, h 5 py. File(ST 2, 'r') as st 2_fid: ST 1 = st 1_fid['/DATA/'+Int. Time+'/Sample. Table']. value ST 2 = st 2_fid['/DATA/'+Int. Time+'/Sample. Table']. value ST 1[ST 1!=0] = 1 ST 2[ST 2!=0] = 1 Diff = (ST 1 - ST 2) ST 1[Diff == 1] = 2 View. Frame(ST 1) HDF & HDF-EOS Workshop XV 17 April 2012

. . and the command line argument parsing. if __name__ == "__main__": import argparse Opt. Parser = argparse. Argument. Parser(description = __doc__) Opt. Parser. add_argument("--ST 1", help="Sample. Table. File 1") Opt. Parser. add_argument("--ST 2", help="Sample. Table. File 2") Opt. Parser. add_argument("--Int. Time", help="Integration Time", default='Long') options = Opt. Parser. parse_args() Compare. ST(options. ST 1, options. ST 2, options. Int. Time) HDF & HDF-EOS Workshop XV 17 April 2012

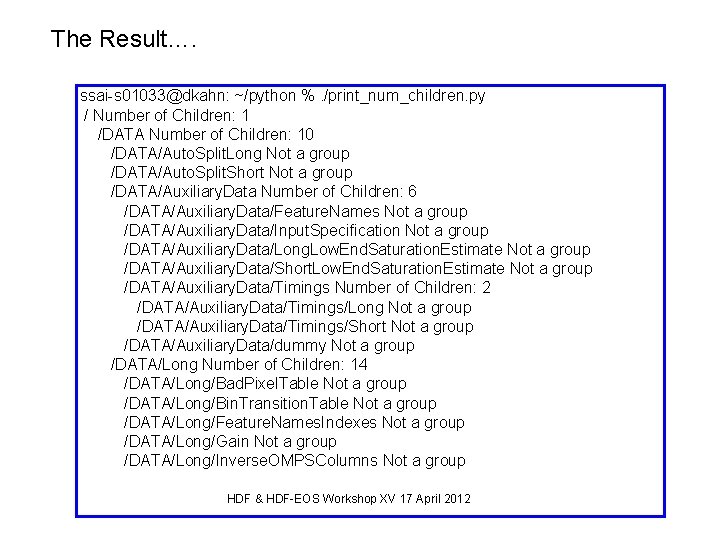
Recursive descent into HDF 5 file Print group names, number of children and dataset names. #!/usr/bin/env python from __future__ import print_function import h 5 py def print_num_children(obj): if isinstance(obj, h 5 py. highlevel. Group): print(obj. name, "Number of Children: ", len(obj)) for Obj. Name in obj: # Obj. Name will a string print_num_children(obj[Obj. Name]) else: print(obj. name, "Not a group") with h 5 py. File('OMPS-NPP-LP_STB', 'r+') as f: print_num_children(f) HDF & HDF-EOS Workshop XV 17 April 2012

The Result…. ssai-s 01033@dkahn: ~/python %. /print_num_children. py / Number of Children: 1 /DATA Number of Children: 10 /DATA/Auto. Split. Long Not a group /DATA/Auto. Split. Short Not a group /DATA/Auxiliary. Data Number of Children: 6 /DATA/Auxiliary. Data/Feature. Names Not a group /DATA/Auxiliary. Data/Input. Specification Not a group /DATA/Auxiliary. Data/Long. Low. End. Saturation. Estimate Not a group /DATA/Auxiliary. Data/Short. Low. End. Saturation. Estimate Not a group /DATA/Auxiliary. Data/Timings Number of Children: 2 /DATA/Auxiliary. Data/Timings/Long Not a group /DATA/Auxiliary. Data/Timings/Short Not a group /DATA/Auxiliary. Data/dummy Not a group /DATA/Long Number of Children: 14 /DATA/Long/Bad. Pixel. Table Not a group /DATA/Long/Bin. Transition. Table Not a group /DATA/Long/Feature. Names. Indexes Not a group /DATA/Long/Gain Not a group /DATA/Long/Inverse. OMPSColumns Not a group HDF & HDF-EOS Workshop XV 17 April 2012

Summary Python with H 5 py and Numpy modules make developing Programs to manipulate HDF 5 files and perform calculations With HDF 5 arrays simpler which increase development speed and reduces errors. HDF & HDF-EOS Workshop XV 17 April 2012