Using Escherichia coli and Saccharomyces cerevisiae as Model















- Slides: 24

Using Escherichia coli and Saccharomyces cerevisiae as Model Systems for Teaching Genomics to Undergraduates Breaking the silence: researching the SIR 2 gene family.

Dr. Myra K. Derbyshire and undergraduate students Mount Saint Mary's College, Emmitsburg, Maryland. Collaborations and affiliations Dr. Jeffery Strathern and Dr. Donald Court NCI-Frederick Cancer Research and Development Center, Frederick, Maryland. GCAT Genome Consortium for Active Teaching Dr. Patrick Brown Stanford Microarray Database Dr. Frederick Blattner University of Wisconsin-Madison E. coli Genome Project.

Yeast SIR 2 is a member of a family of genes conserved from E. coli to humans.

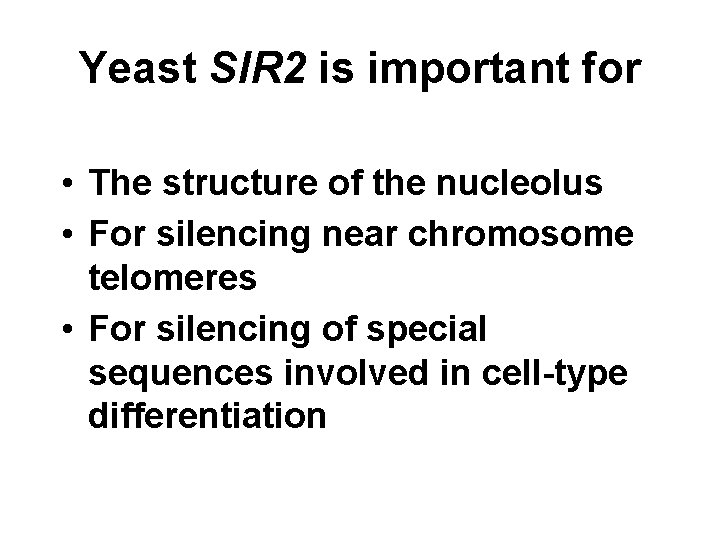
Yeast SIR 2 is important for • The structure of the nucleolus • For silencing near chromosome telomeres • For silencing of special sequences involved in cell-type differentiation

Silencing : • The term given to a mechanism by which gene expression in regions of the genome is repressed • Silencing can be modified by changes in chromatin structure

• In eukaryotes, altering chromatin states around a gene allows cells to achieve complex patterns of regulation. • SIR 2 p is a component of chromatin • SIR 2 p sets up silent chromatin states around the genes it regulates.

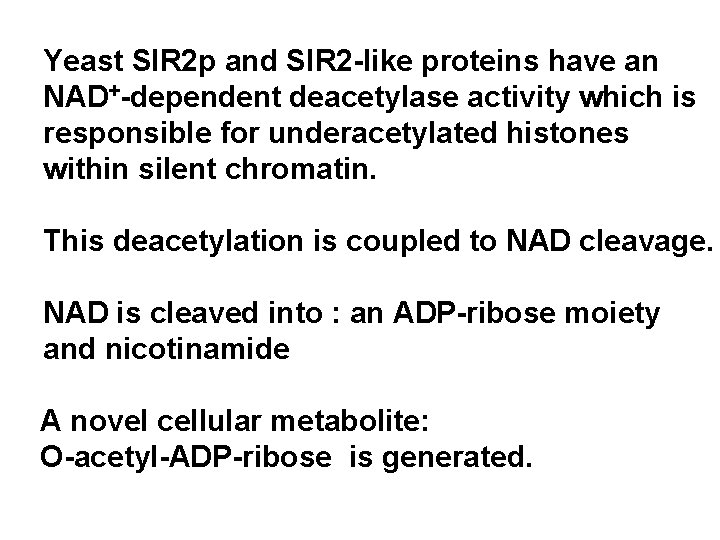
Yeast SIR 2 p and SIR 2 -like proteins have an NAD+-dependent deacetylase activity which is responsible for underacetylated histones within silent chromatin. This deacetylation is coupled to NAD cleavage. NAD is cleaved into : an ADP-ribose moiety and nicotinamide A novel cellular metabolite: O-acetyl-ADP-ribose is generated.

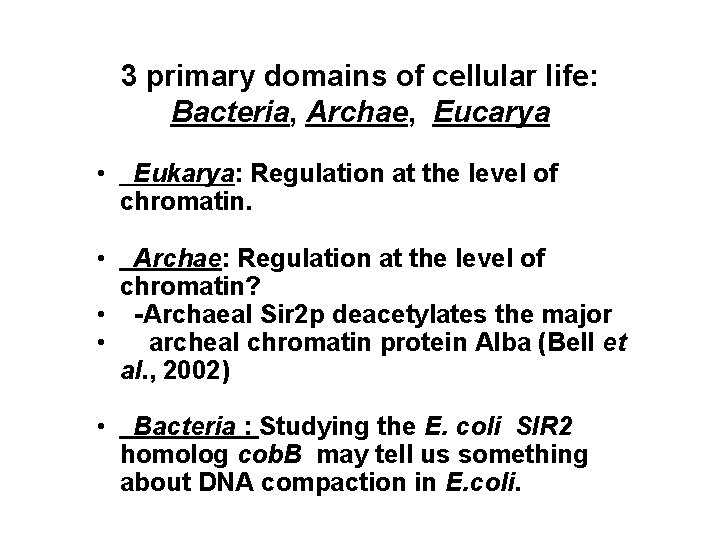
3 primary domains of cellular life: Bacteria, Archae, Eucarya • Eukarya: Regulation at the level of chromatin. • Archae: Regulation at the level of chromatin? • -Archaeal Sir 2 p deacetylates the major • archeal chromatin protein Alba (Bell et al. , 2002) • Bacteria : Studying the E. coli SIR 2 homolog cob. B may tell us something about DNA compaction in E. coli.

There are five members of the SIR 2 gene family in Saccharomyces cerevisiae (SIR 2, HST 1 -HST 4) – Little is known of the roles of the HST 1 -HST 4 genes. – HST 1 -HST 4 gene products may modify chromatin structure at different chromosomal locations than those regulated by SIR 2 p.

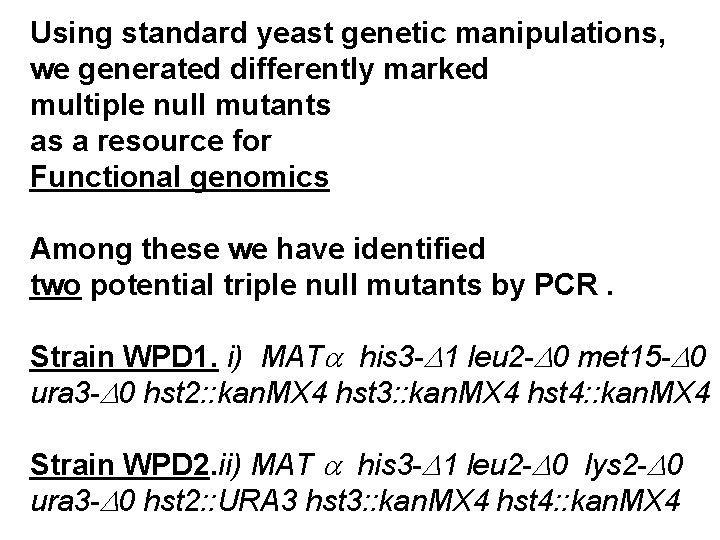
Using standard yeast genetic manipulations, we generated differently marked multiple null mutants as a resource for Functional genomics Among these we have identified two potential triple null mutants by PCR. Strain WPD 1. i) MATa his 3 -D 1 leu 2 -D 0 met 15 -D 0 ura 3 -D 0 hst 2: : kan. MX 4 hst 3: : kan. MX 4 hst 4: : kan. MX 4 Strain WPD 2. ii) MAT a his 3 -D 1 leu 2 -D 0 lys 2 -D 0 ura 3 -D 0 hst 2: : URA 3 hst 3: : kan. MX 4 hst 4: : kan. MX 4

Students are currently focusing on microarray analysis of a hst 3: : kan. MX 4 hst 4: : kan. MX 4 double mutant generated as follows Cross: Research Genetics strain #3550: MATa his 3 - 1 leu 2 - 0 met 15 - 0 ura 3 - 0 hst 4: : kan. MX 4 X Research Genetics strain #11801: MAT his 3 - 1 leu 2 - 0 lys 2 - 0 ura 3 - 0 hst 3: : kan. MX 4

The resulting diploid was sporulated, tetrads were dissected and spore clones were screened for the hst 3: : kan. MX 4 hst 4: : kan. MX 4 double mutant by Southern analysis.

• Summer 2000. MSM undergraduates generated an E. coli cob. B null mutant • Using a defective prophage mediated recombination system described by (Yu et al 2000) • U. W. Madison E. coli genome project E. coli mutant strains are available for functional analysis.

Milieu • Liberal Arts College situated close to Frederick Maryland • College missions include: teaching, scholarship and community service. • Research for 20 years post Ph. D. Full time teaching last 4 years. • Support from NCI-FCRDC and GCAT.

Number of students in MSM Genomics initiative Fall 2000: Genetics (11) Spring 2001: Research (1) Summer 2001. Research (3) Fall 2001: Genetics (18) Summer 2002. Research (2) Fall 2002: Genetics (25 ? ) Research (4)

MSM students through genomic research Strengthen - conceptual understanding - problem solving skills Have access to the virtual learning /teaching/research/service community of GCAT. Immerse themselves through Genomics in different model genetic systems.

Global Goals of Genomics: – To assess an organisms genetic complement. – To determine which genes are expressed and under what conditions. – To determine the function of protein products.

Students in their study of the SIR 2 family: predict gene function Identify SIR 2 family homologs – Interact with Gen. Bank (database of all publicly available DNA and derived protein sequences) – Carry out data base searches to identify family members using: PSI-Blast and know motifs.

Students in their study of the SIR 2 family appreciate : – multiple sequence alignments. – Structural similarity does not imply common function. – To predict function one needs experimental proof of function

MSM students studying the SIR 2 family Compared gene expression patterns of two meiotic segregants (HST 3 HST 4 and hst 3 hst 4) using microarray technology (GCAT) This data is viewable on a public search of the Stanford Microarray database.

Undergraduate access to Yeast Microarray technology was made possible through affiliation with GCAT/Stanford Microarray database. MSM Undergraduates are: – Repeating this microarray comparison using three independent RNA isolations – They will analyze data from multiple experiments and correlate patterns of gene expression

Planned student microarray experiment: made possible through affiliation with GCAT/ UW-Madison E. coli genome project Gene expression patterns of an E. coli mutant having a null mutation in the cob. B gene and an isogenic wild-type strain

Feedback from some MSM students who participated in the collaboration • – – – I am working on protein structure predictions algorithms…very neat stuff! (Pursuing a Ph. D. in Bioinformatics). Thank you again for the wonderful opportunity and experience you afforded me in genetics class (Pursuing a graduate program in Cellular and Molecular medicine) Thank you for everything you have given and taught me. I carry it with me all the time. (Pursuing a Masters in Bioscience and technology).

Collaborations and affiliations Dr. Jeffery Strathern and Dr. Donald Court NCI-Frederick Cancer Research and Development Center, Frederick, Maryland. GCAT Genome Consortium for Active Teaching Dr. Patrick Brown Stanford Microarray Database Dr. Frederick Blattner University of Wisconsin-Madison E. coli Genome Project.