Using Definitive Screening Design to Optimise Recovery PreTreatment
- Slides: 3

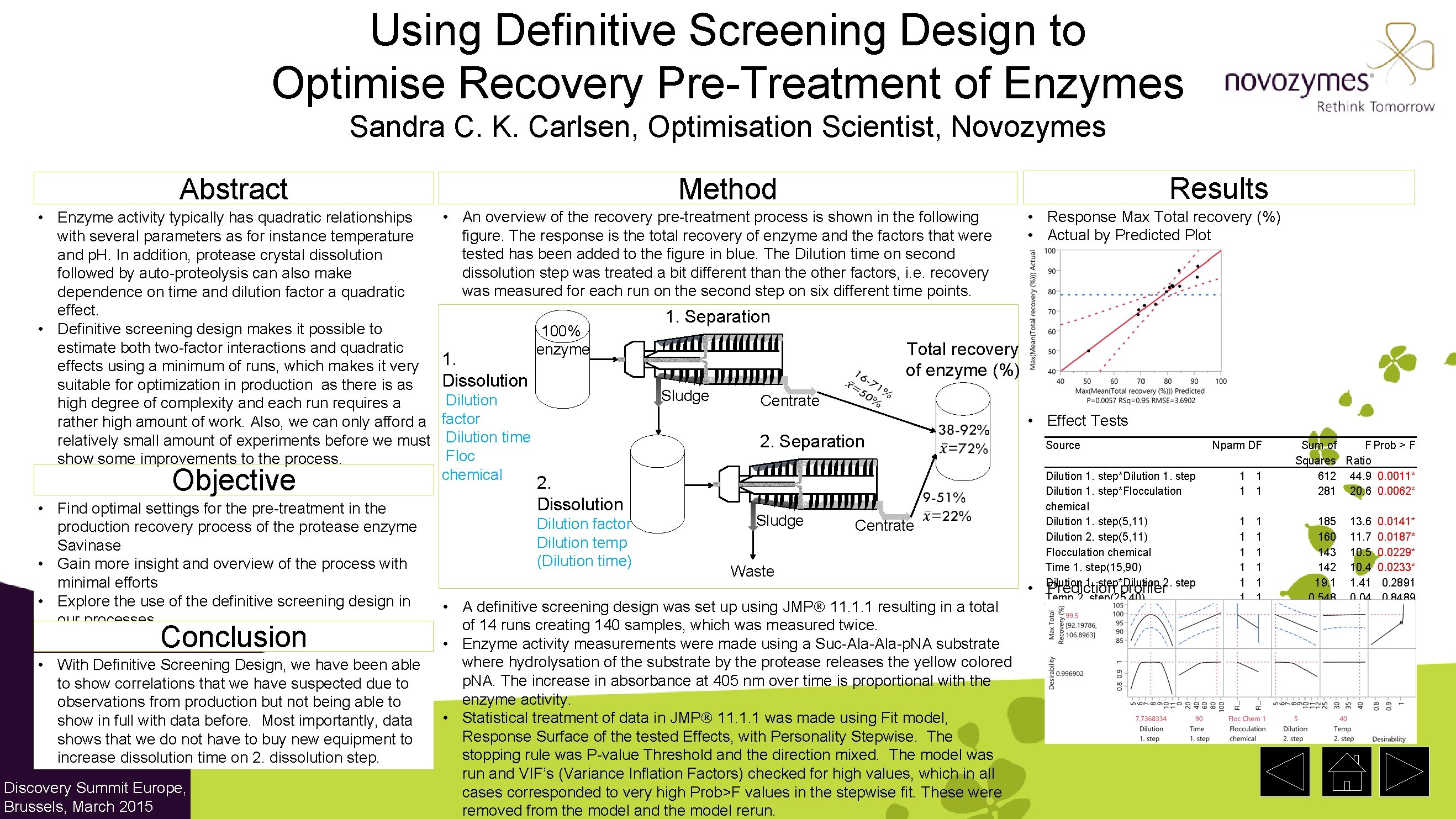
Using Definitive Screening Design to Optimise Recovery Pre-Treatment of Enzymes Sandra C. K. Carlsen, Optimisation Scientist, Novozymes • Enzyme activity typically has quadratic relationships with several parameters as for instance temperature and p. H. In addition, protease crystal dissolution followed by auto-proteolysis can also make dependence on time and dilution factor a quadratic effect. • Definitive screening design makes it possible to estimate both two-factor interactions and quadratic effects using a minimum of runs, which makes it very suitable for optimization in production as there is as high degree of complexity and each run requires a rather high amount of work. Also, we can only afford a relatively small amount of experiments before we must show some improvements to the process. Objective • Find optimal settings for the pre-treatment in the production recovery process of the protease enzyme Savinase • Gain more insight and overview of the process with minimal efforts • Explore the use of the definitive screening design in our processes Conclusion • With Definitive Screening Design, we have been able to show correlations that we have suspected due to observations from production but not being able to show in full with data before. Most importantly, data shows that we do not have to buy new equipment to increase dissolution time on 2. dissolution step. Discovery Summit Europe, Brussels, March 2015 Results Method Abstract • An overview of the recovery pre-treatment process is shown in the following figure. The response is the total recovery of enzyme and the factors that were tested has been added to the figure in blue. The Dilution time on second dissolution step was treated a bit different than the other factors, i. e. recovery was measured for each run on the second step on six different time points. 1. Dissolution Dilution factor Dilution time Floc chemical 100% enzyme 1. Separation Total recovery of enzyme (%) Sludge Centrate 2. Separation 2. Dissolution Dilution factor Dilution temp (Dilution time) • Response Max Total recovery (%) • Actual by Predicted Plot • Effect Tests Source Sludge Centrate Waste • A definitive screening design was set up using JMP 11. 1. 1 resulting in a total of 14 runs creating 140 samples, which was measured twice. • Enzyme activity measurements were made using a Suc-Ala-p. NA substrate where hydrolysation of the substrate by the protease releases the yellow colored p. NA. The increase in absorbance at 405 nm over time is proportional with the enzyme activity. • Statistical treatment of data in JMP 11. 1. 1 was made using Fit model, Response Surface of the tested Effects, with Personality Stepwise. The stopping rule was P-value Threshold and the direction mixed. The model was run and VIF’s (Variance Inflation Factors) checked for high values, which in all cases corresponded to very high Prob>F values in the stepwise fit. These were removed from the model and the model rerun. • Dilution 1. step*Dilution 1. step*Flocculation chemical Dilution 1. step(5, 11) Dilution 2. step(5, 11) Flocculation chemical Time 1. step(15, 90) Dilution 1. step*Dilution 2. step Prediction profiler Temp 2. step(25, 40) Nparm DF 1 1 1 1 Sum of F Prob > F Squares Ratio 612 44. 9 0. 0011* 281 20. 6 0. 0062* 185 160 143 142 19. 1 0. 548 13. 6 11. 7 10. 5 10. 4 1. 41 0. 04 0. 0141* 0. 0187* 0. 0229* 0. 0233* 0. 2891 0. 8489

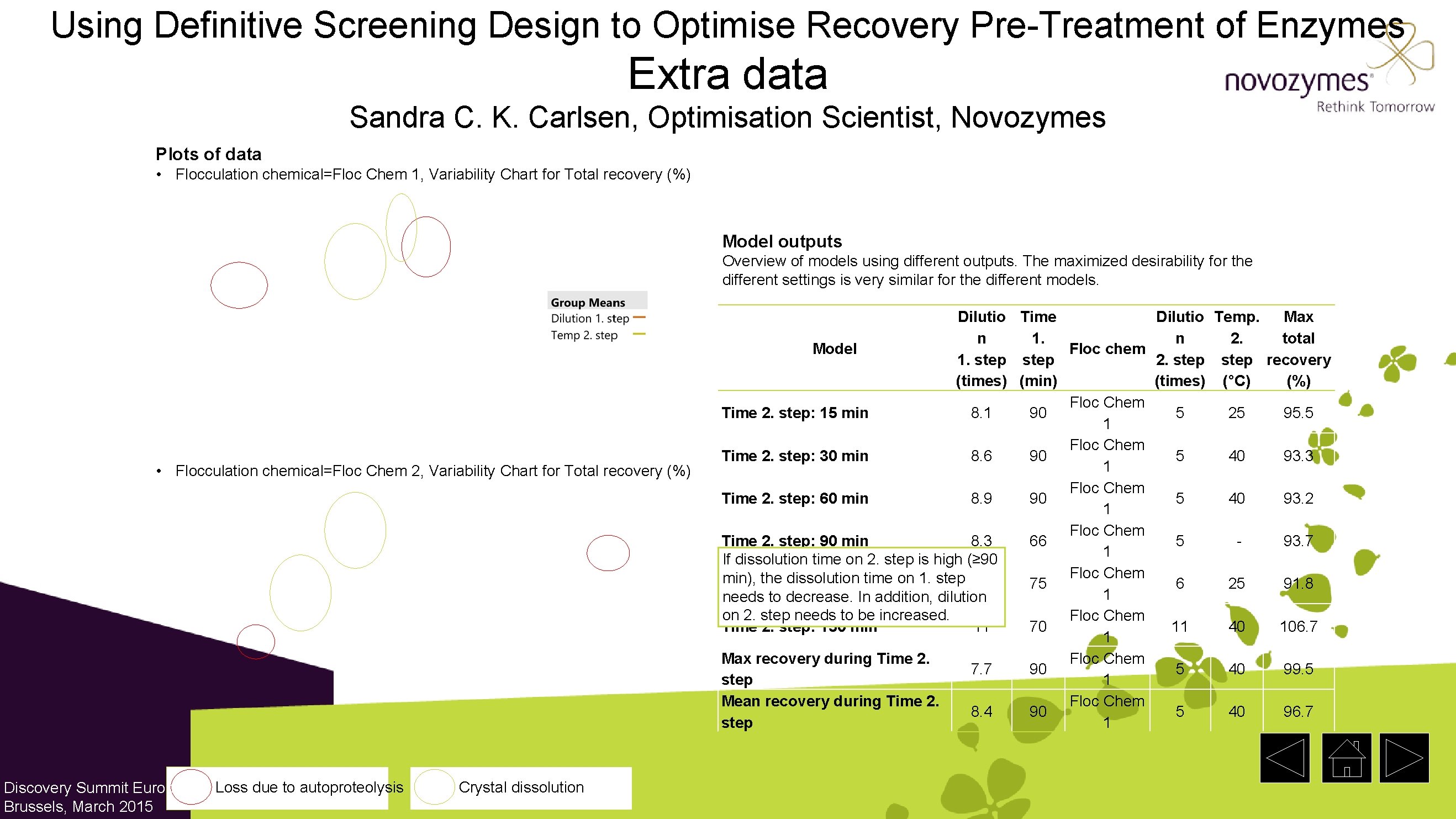
Using Definitive Screening Design to Optimise Recovery Pre-Treatment of Enzymes Extra data Sandra C. K. Carlsen, Optimisation Scientist, Novozymes Plots of data • Flocculation chemical=Floc Chem 1, Variability Chart for Total recovery (%) Model outputs Overview of models using different outputs. The maximized desirability for the different settings is very similar for the different models. • Flocculation chemical=Floc Chem 2, Variability Chart for Total recovery (%) Discovery Summit Europe, Brussels, March 2015 Loss due to autoproteolysis Crystal dissolution Dilutio Time Dilutio Temp. Max n 1. n 2. total Model Floc chem 1. step 2. step recovery (times) (min) (times) (°C) (%) Floc Chem Time 2. step: 15 min 8. 1 90 5 25 95. 5 1 Floc Chem Time 2. step: 30 min 8. 6 90 5 40 93. 3 1 Floc Chem Time 2. step: 60 min 8. 9 90 5 40 93. 2 1 Floc Chem Time 2. step: 90 min 8. 3 66 5 93. 7 1 If dissolution time on 2. step is high (≥ 90 Floc Chem min), the dissolution time on 1. step Time 2. step: 120 min 8. 6 75 6 25 91. 8 1 needs to decrease. In addition, dilution on 2. step needs to be increased. Floc Chem Time 2. step: 150 min 11 70 11 40 106. 7 1 Max recovery during Time 2. Floc Chem 7. 7 90 5 40 99. 5 step 1 Mean recovery during Time 2. Floc Chem 8. 4 90 5 40 96. 7 step 1

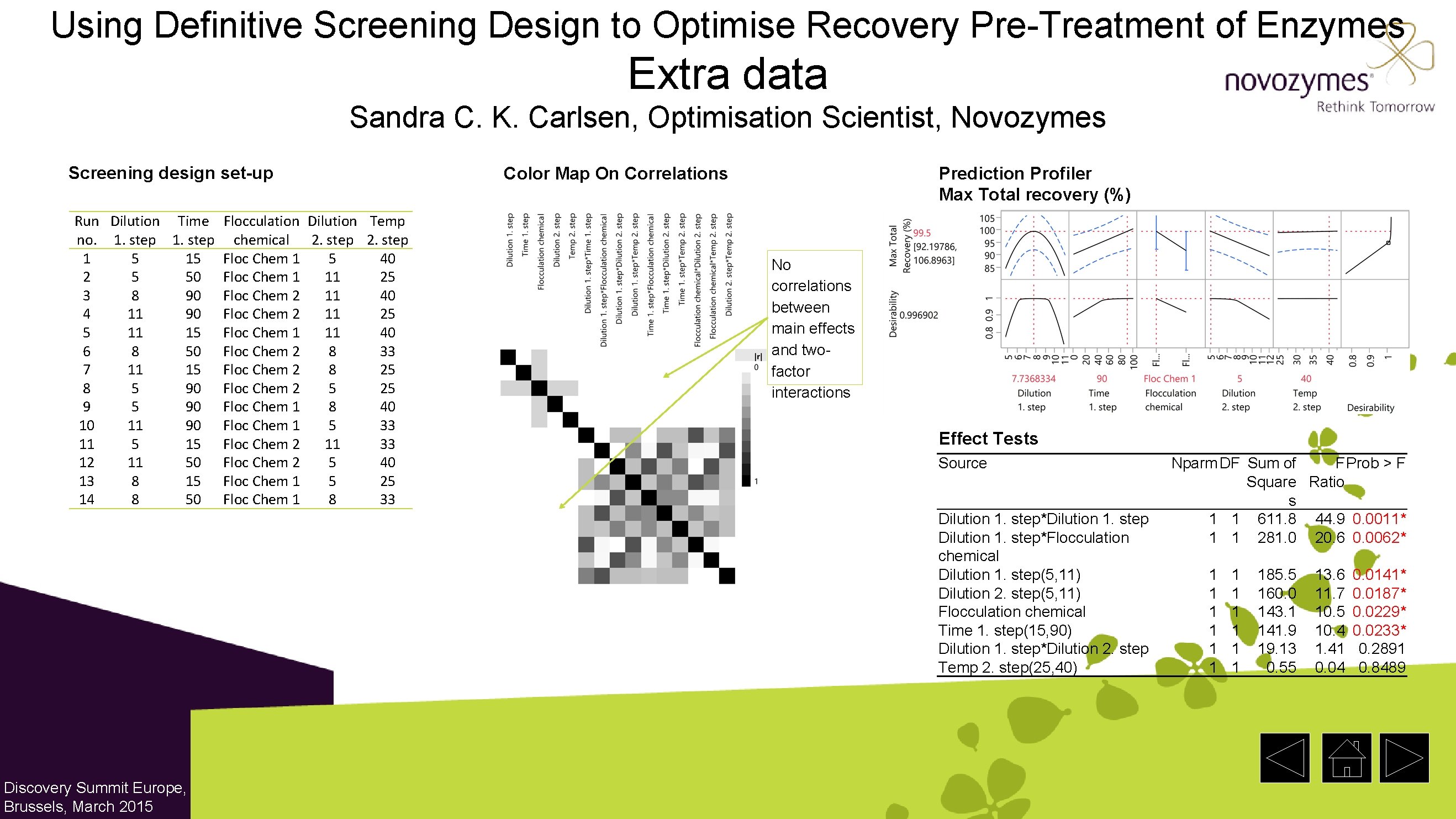
Using Definitive Screening Design to Optimise Recovery Pre-Treatment of Enzymes Extra data Sandra C. K. Carlsen, Optimisation Scientist, Novozymes Screening design set-up Run Dilution Time Flocculation Dilution Temp no. 1. step chemical 2. step 1 5 15 Floc Chem 1 5 40 2 5 50 Floc Chem 1 11 25 3 8 90 Floc Chem 2 11 40 4 11 90 Floc Chem 2 11 25 5 11 15 Floc Chem 1 11 40 6 8 50 Floc Chem 2 8 33 7 11 15 Floc Chem 2 8 25 8 5 90 Floc Chem 2 5 25 9 5 90 Floc Chem 1 8 40 10 11 90 Floc Chem 1 5 33 11 5 15 Floc Chem 2 11 33 12 11 50 Floc Chem 2 5 40 13 8 15 Floc Chem 1 5 25 14 8 50 Floc Chem 1 8 33 Color Map On Correlations Prediction Profiler Max Total recovery (%) No correlations between main effects and twofactor interactions Effect Tests Source Dilution 1. step*Dilution 1. step*Flocculation chemical Dilution 1. step(5, 11) Dilution 2. step(5, 11) Flocculation chemical Time 1. step(15, 90) Dilution 1. step*Dilution 2. step Temp 2. step(25, 40) Discovery Summit Europe, Brussels, March 2015 Nparm DF Sum of F Prob > F Square Ratio s 1 1 611. 8 44. 9 0. 0011* 1 1 281. 0 20. 6 0. 0062* 1 1 1 185. 5 160. 0 143. 1 141. 9 19. 13 0. 55 13. 6 11. 7 10. 5 10. 4 1. 41 0. 04 0. 0141* 0. 0187* 0. 0229* 0. 0233* 0. 2891 0. 8489