Using BLAST to Identify Species from Proteins Adapted
Using BLAST to Identify Species from Proteins Adapted from College Board’s “Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST”

What we know so far. . . ● We already know that DNA can be used to identify animal species at a genetic level ● According to the Central Dogma of Biology, DNA is transcribed into RNA and translated into proteins ● If DNA is not present in a bone specimen, other biological material can be acquired to identify a fossil

Today’s Goal ● Become acquainted with BLAST for protein analysis ● Identify the relation of extinct dinosaurs to modern species and where dinosaurs fit in on a family tree ● Understand how phylogenetic trees depict evolutionary relationships

What you will need to know ● What is BLAST, and how is it used? ● Why is BLAST important? ● What is a phylogenetic tree? ● What are orthologs? ● How can proteins be used to identify evolutionary relationships?

What is BLAST? BLAST Basic Local Alignment Search Tool

Why is BLAST useful? ● BLAST makes it easy to search databases that contain many millions of partial, full, and potential gene sequences ● It can be used to compare sequences from one species to another to identify similarities or differences ● Knowing the identity of gene sequences from many species allows scientists to track evolutionary heritage of those species
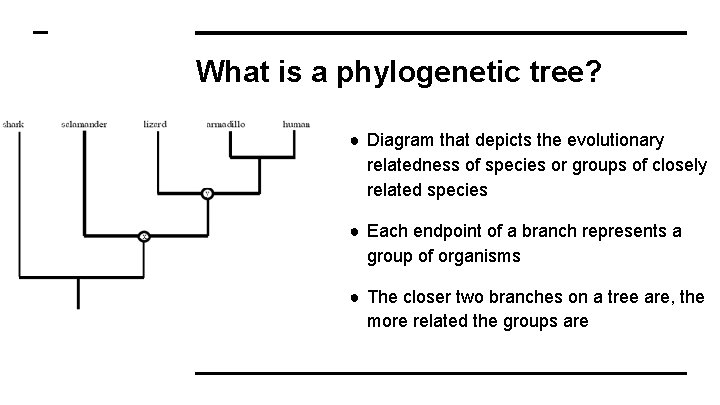
What is a phylogenetic tree? ● Diagram that depicts the evolutionary relatedness of species or groups of closely related species ● Each endpoint of a branch represents a group of organisms ● The closer two branches on a tree are, the more related the groups are
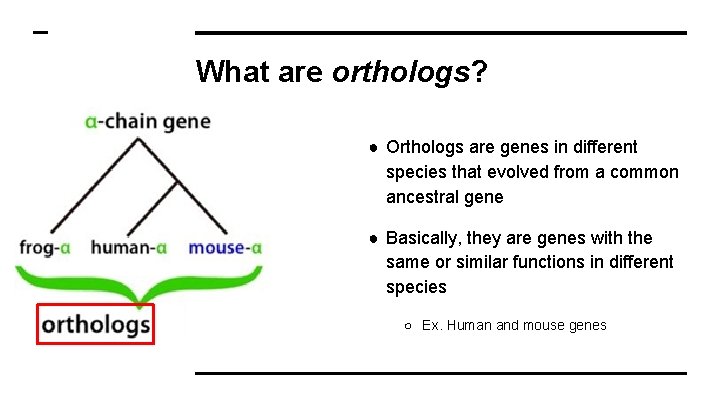
What are orthologs? ● Orthologs are genes in different species that evolved from a common ancestral gene ● Basically, they are genes with the same or similar functions in different species ○ Ex. Human and mouse genes
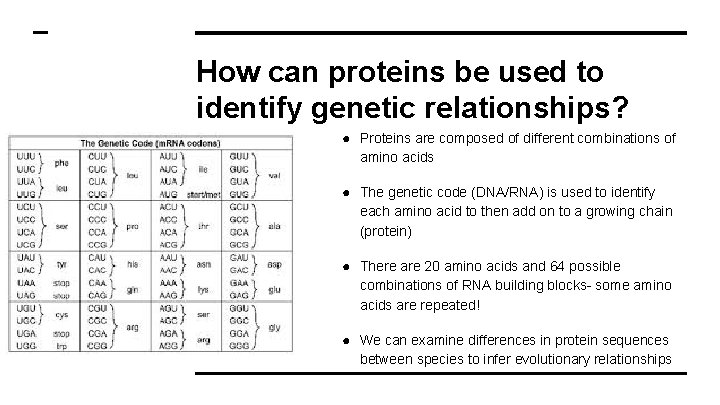
How can proteins be used to identify genetic relationships? ● Proteins are composed of different combinations of amino acids ● The genetic code (DNA/RNA) is used to identify each amino acid to then add on to a growing chain (protein) ● There are 20 amino acids and 64 possible combinations of RNA building blocks- some amino acids are repeated! ● We can examine differences in protein sequences between species to infer evolutionary relationships

Activity: Identify Evolutionarily Related Protein Sequences ● You are a member of a scientific team that has discovered 3 well preserved bone fossil specimens ○ T. rex, mastodon, and hadrosaur ● Small amounts of tissue have been removed from the fossil ○ Unusual in such ancient specimens ● Amino acid sequences have been extracted from protein fragments
Activity: Identify Evolutionarily Related Protein Sequences ● Use BLAST to compare these amino acid sequences to protein sequences from other species ● Then, create a phylogenetic tree to explore where these extinct species might branch off from the evolutionary tree
Step 1 ● Form an initial hypothesis as to where and why you think the three extinct species (T. rex, hadrosaur, mastodon) belong on the phylogenetic tree

Step 2 ● Locate and download the protein fragment files for the fossilized bone specimens

Step 3 ● Upload the sequence into BLAST ○ Go to BLAST homepage: https: //blast. ncbi. nlm. nih. gov/Bla st. cgi ○ Click on “Protein BLAST”
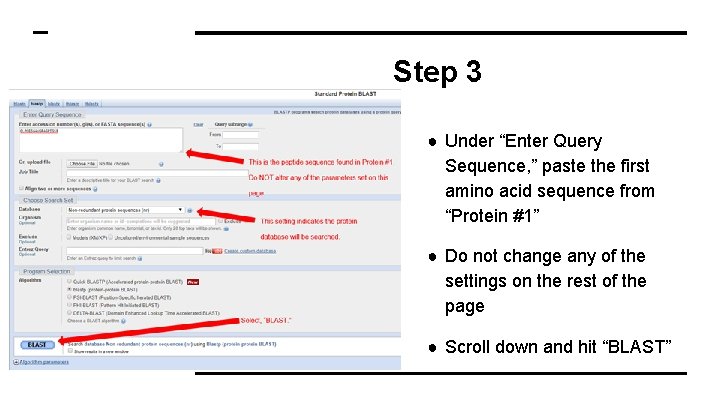
Step 3 ● Under “Enter Query Sequence, ” paste the first amino acid sequence from “Protein #1” ● Do not change any of the settings on the rest of the page ● Scroll down and hit “BLAST”
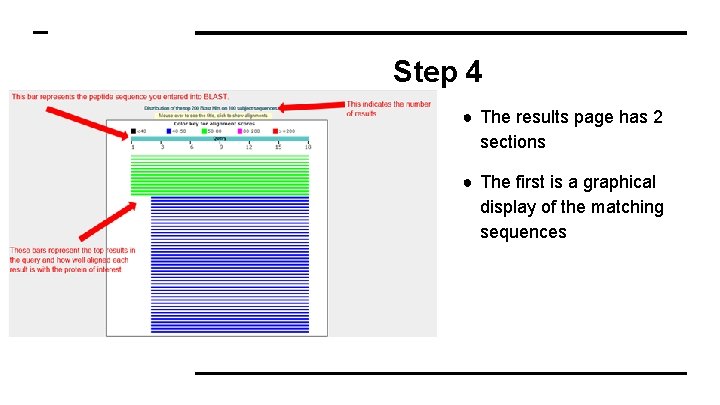
Step 4 ● The results page has 2 sections ● The first is a graphical display of the matching sequences

Step 4 ● Scroll down to the section titled “Sequences Producing Significant Alignment” ● The species in this list are the ones with sequences identical to or most similar to the amino acid sequence of interest ● The most similar sequence is listed first ● If you click on a result listed, you will get a full report of the species
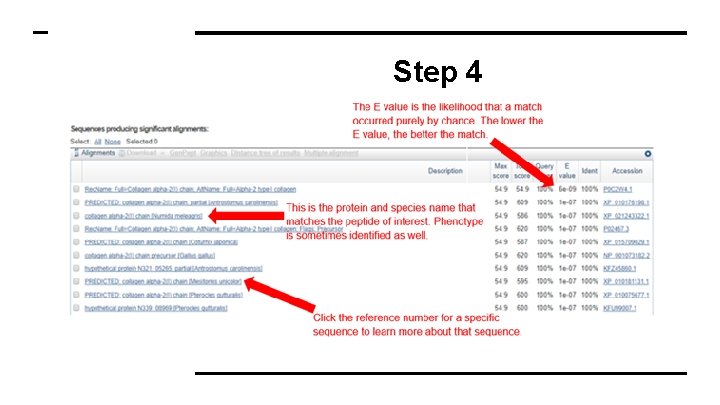
Step 4
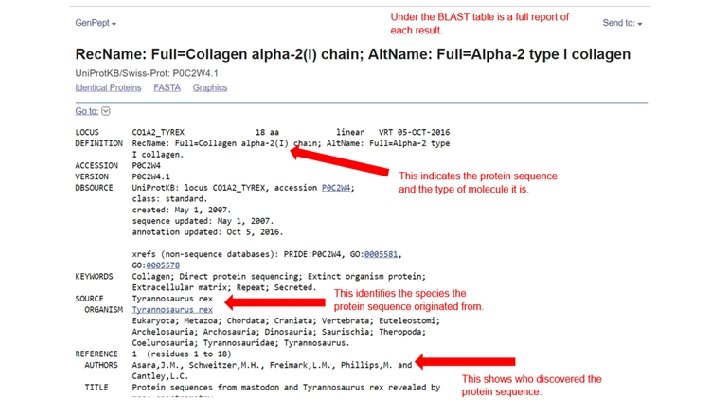

Now that you have this information. . . ● Follow the PDF instructions for the BLAST search of the fossilized T. rex bone amino acid sequence ● If you still have time at the end of class and want to practice and explore more, repeat the activity for the hadrosaur and mastodon fossil specimens
- Slides: 20