Unix tools Regular expressions grep sed AWK Regular




















- Slides: 33

Unix tools • Regular expressions • grep • sed • AWK

Regular expressions • Sequence of characters that define a search pattern • banana matches the text banana • b[A-Z 0 -9. _%+-]+@[A-Z 0 -9. -]+. [A-Z]{2, }b matches email addresses • Easier to write than read. . .

grep (globally search a regular expression and print) • grep ‘>’ sequence. fasta • prints all lines containing ‘>’ in sequence. fasta • grep -c ‘b[A-Z 0 -9. _%+-]+@[A-Z 0 -9. -]+. [A-Z]{2, }b‘ things. txt • prints number of lines containing email addresses in things. txt • examples

Regular expressions in Python • https: //docs. python. org/2/library/re. html • Python language vs. regex language • examples

sed (stream editor) • makes changes in a file • s for substitution • sed ‘s/day/night/’ old > new changes first occurrence of day on each line in old to night in new • examples • http: //www. grymoire. com/Unix/Sed. html#uh-64

AWK • data extraction and reporting • pattern { action } • pattern specifies a test that is performed with each line read as input • useful for processing tables of data • examples • http: //www. grymoire. com/Unix/Awk. html#uh-0

Most bioinformatics coursework focuses on algorithms, with perhaps some components devoted to learning programming skills and learning how to use existing bioinformatics software. Unfortunately, for students who are preparing for a research career, this type of curriculum fails to address many of the day-to-day organizational challenges associated with performing computational experiments…I will focus on relatively mundane issues such as organizing files and directories and documenting progress. These issues are important because poor organizational choices can lead to significantly slower research progress.

Principle 1: Someone unfamiliar with your project should be able to look at your computer files and understand in detail what you did and why. • That someone might be • • • a reader of your article a collaborator a future labmate your advisor you, months later. Principle 2: Everything you do, you will probably have to do over again. • Reanalysis with different algorithm or data


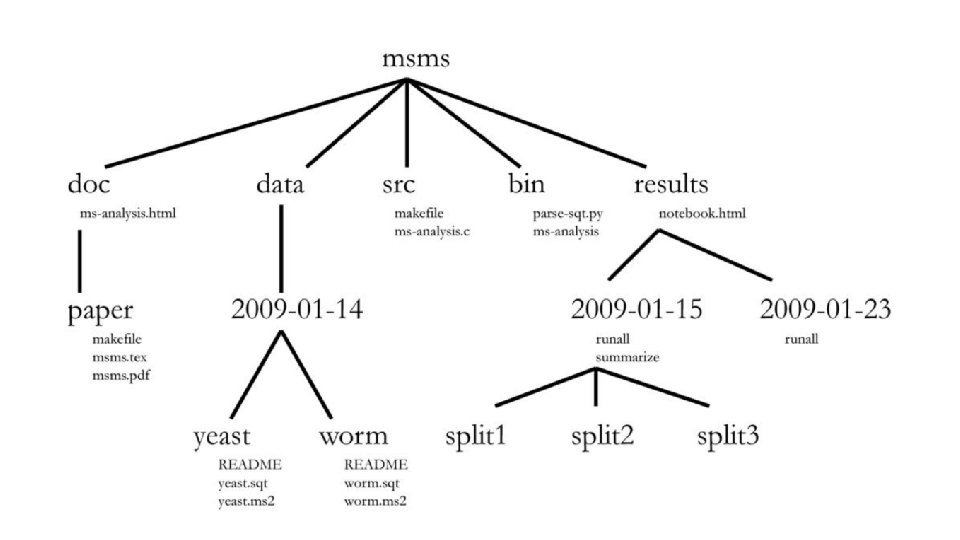
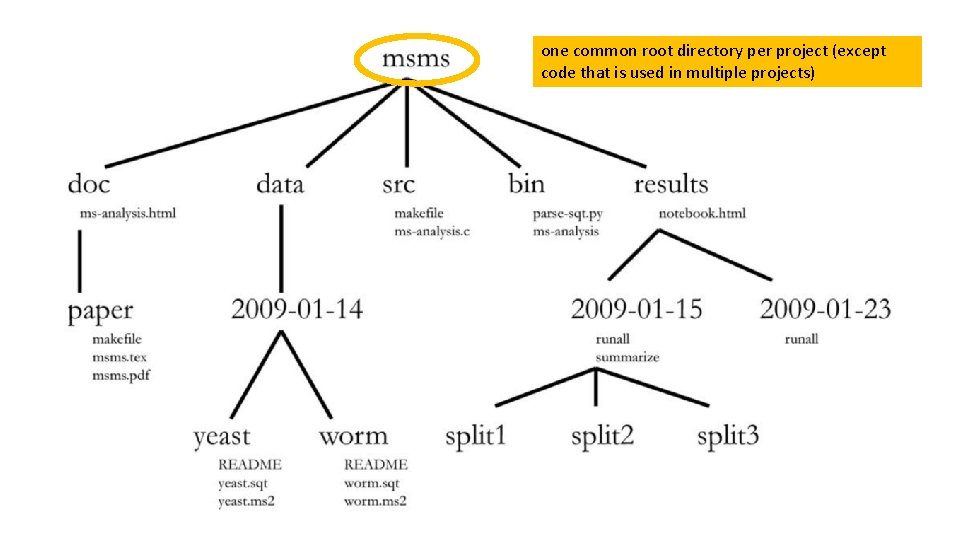
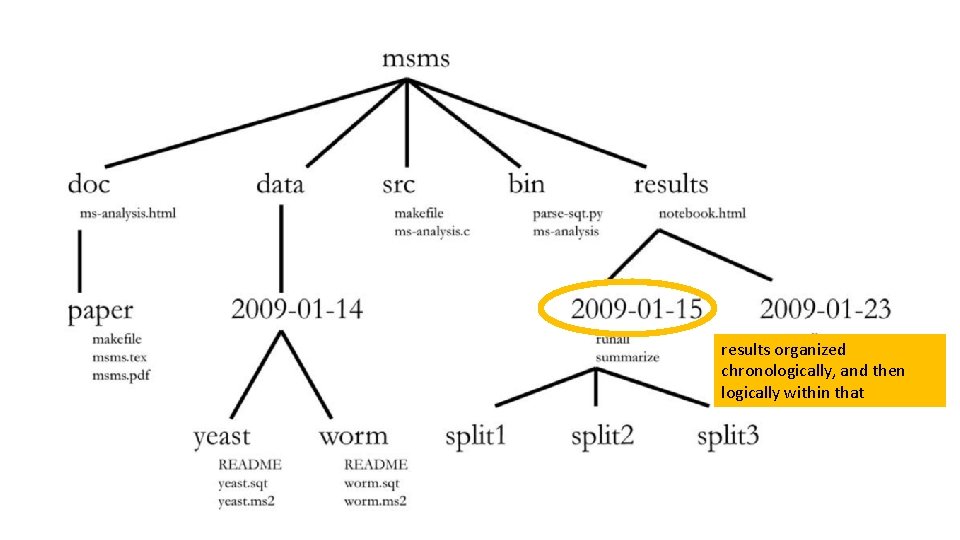
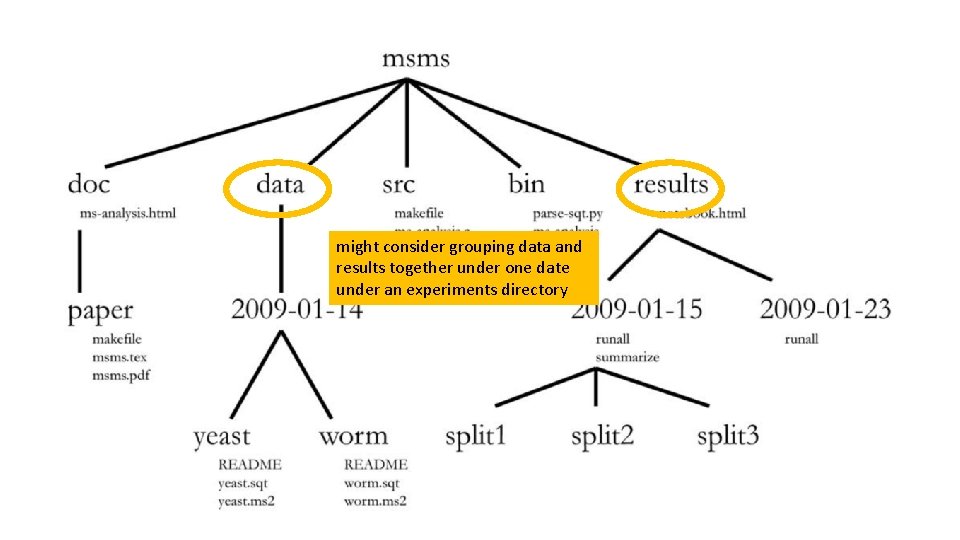
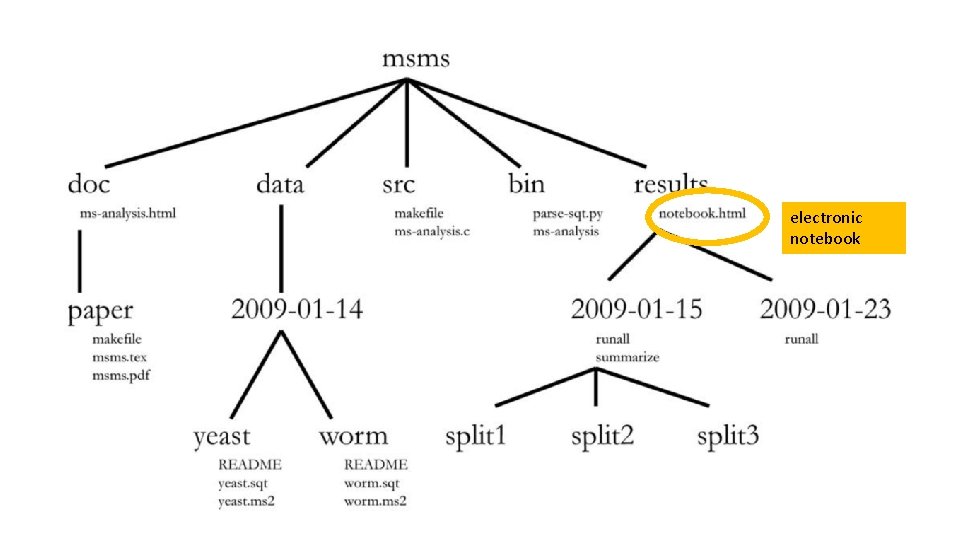
one common root directory per project (except code that is used in multiple projects)

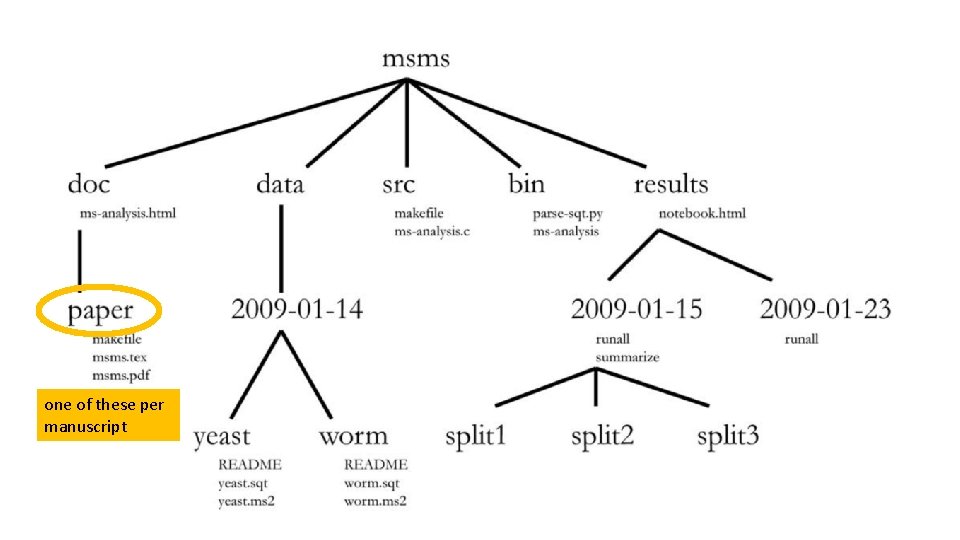
one of these per manuscript

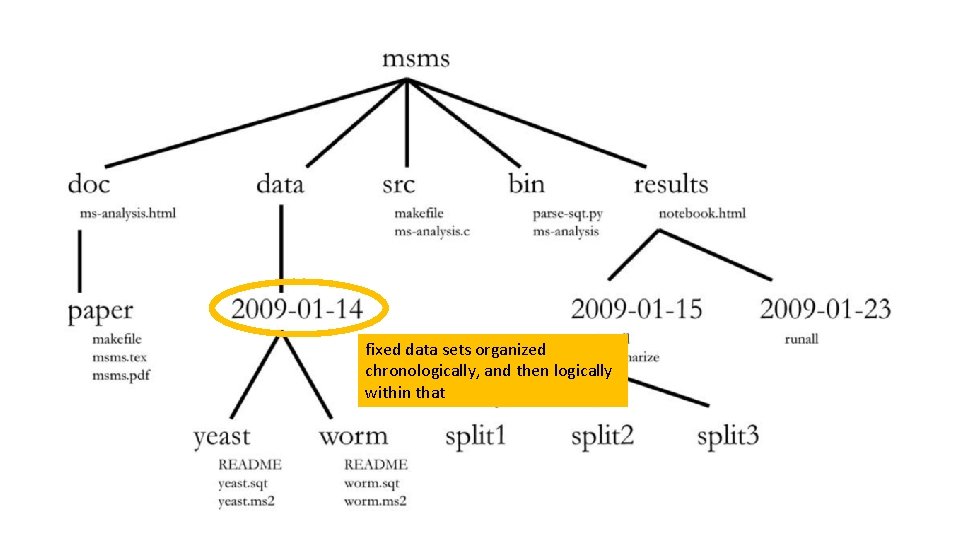
fixed data sets organized chronologically, and then logically within that

results organized chronologically, and then logically within that

might consider grouping data and results together under one date under an experiments directory

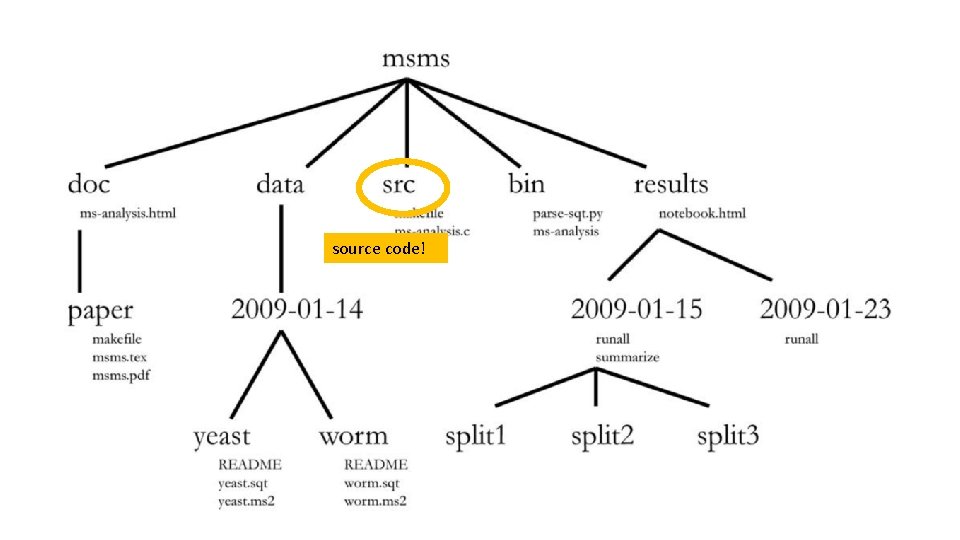
source code!

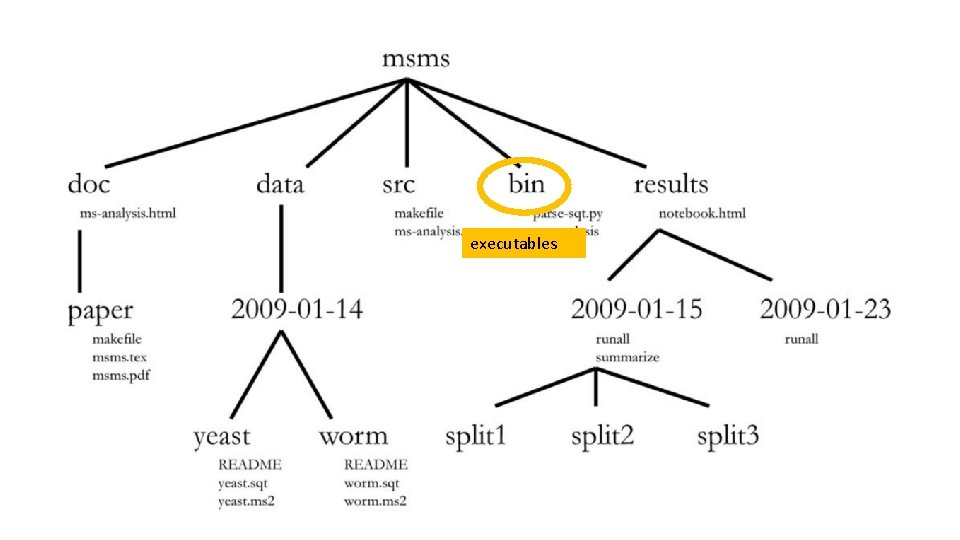
executables

electronic notebook

Lab notebook • relatively verbose • links, images, tables, plots • observations, conclusions, how you got there, ideas for future • document failed experiments well, too • conversations, emails • options: • evernote • onenote • whatever you like…

Carrying out a single experiment 1. Record every operation you perform • README • driver script (runall) • parallel to lab notebook entry—gory details here, prose description in notebook • organization depends somewhat on what tools you’re using • one big R script vs. different pieces of compiled code

Carrying out a single experiment 2. Comment generously • should be understandable what you’re doing solely from the comments 3. Avoid editing intermediate files by hand • want script to be completely automatic • include the sed, awk, grep commands, etc. 4. Store all file and directory names in the script • easier to keep track of and modify if they’re all in one place

Carrying out a single experiment 5. Use relative pathnames • so that it can work for other people who check it out 6. Make the script restartable • if (output file does not exist) then perform operation • allow your script to let you skip rerunning long steps if unnecessary • progress output

Carrying out a single experiment • One script to run the experiment (runall) • final line calls summarize • One script to summarize the results (summarize) • creates plots, tables, or other summary • can interpret partially completed experiment

Handling and preventing errors 1. Write robust code to detect errors • Check validity of parameters, other inputs • Existing programs to read standard file formats 2. When an error occurs, abort • Print message to standard error • Exit with nonzero exit status 3. Create each output file with a temporary name, rename after complete • Prevents partial results from being mistaken for full results

Command lines vs. scripts vs. programs • How much effort to put into software engineering? • quick set of scripts hacked together • over-engineered automation • something in the middle • Iterative improvement of scripts • one script eventually broken into many • change of programming language

Command lines vs. scripts vs. programs • Categories of scripts: • Driver • One or two per project • Single-use • e. g. converting some arbitrary file format in an experiment • Project-specific • used by multiple experiments within the project • Multi-project • e. g. dealing with common file formats, generating common plots

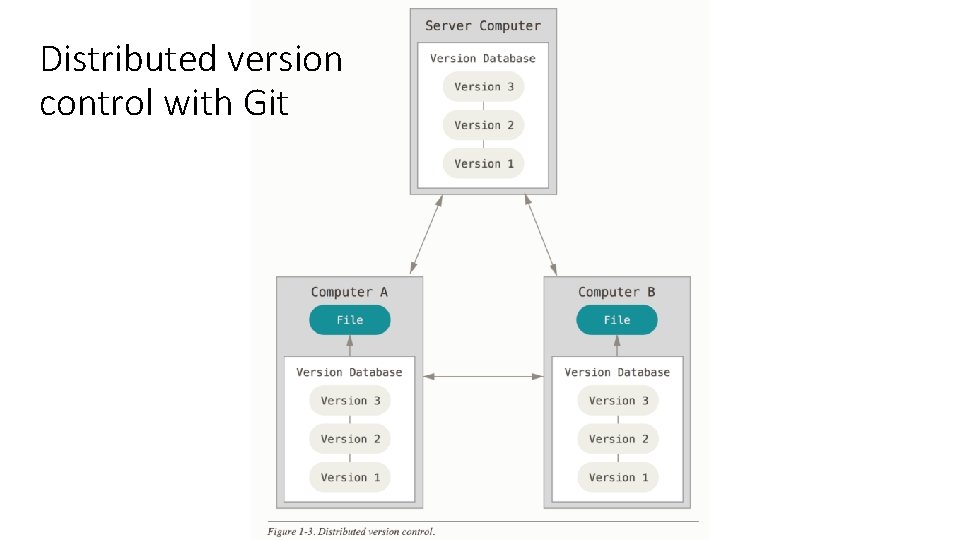
The value of version control • backups • you might not backup things on your local machine regularly • easier to retrieve previous versions than through system administrator • historical record • programs evolve over the course of the project • simpler than dealing with a bunch of different copies of the file • reproduce an experiment with code from some specific date • collaboration • edit same file simultaneously • merge later

The value of version control • requires discipline • check in at least once a day • if your code is currently broken, can check into a “branch” and then merge into main project (“trunk”) later • should only be used for files you edit by hand • no data, compiled programs, results • can tell version control system to ignore certain types of files

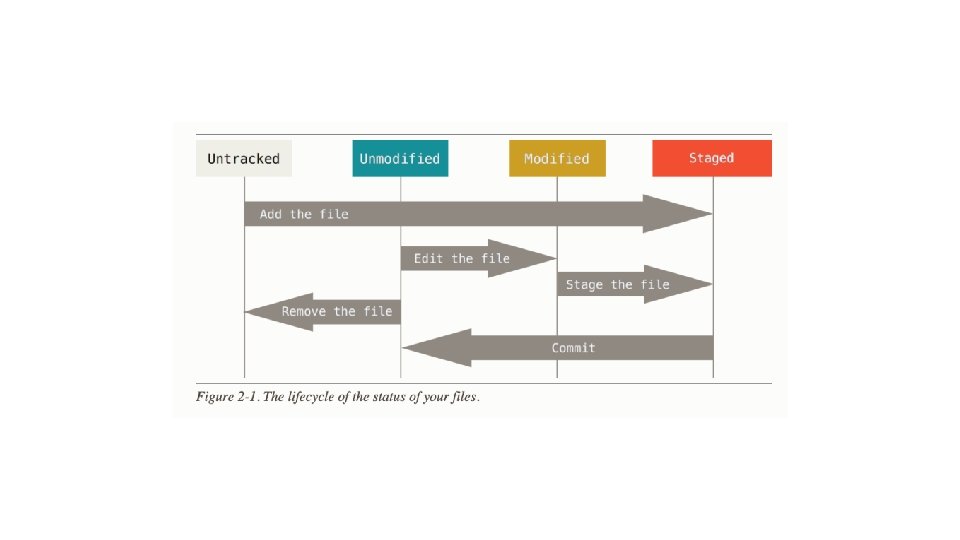
Distributed version control with Git

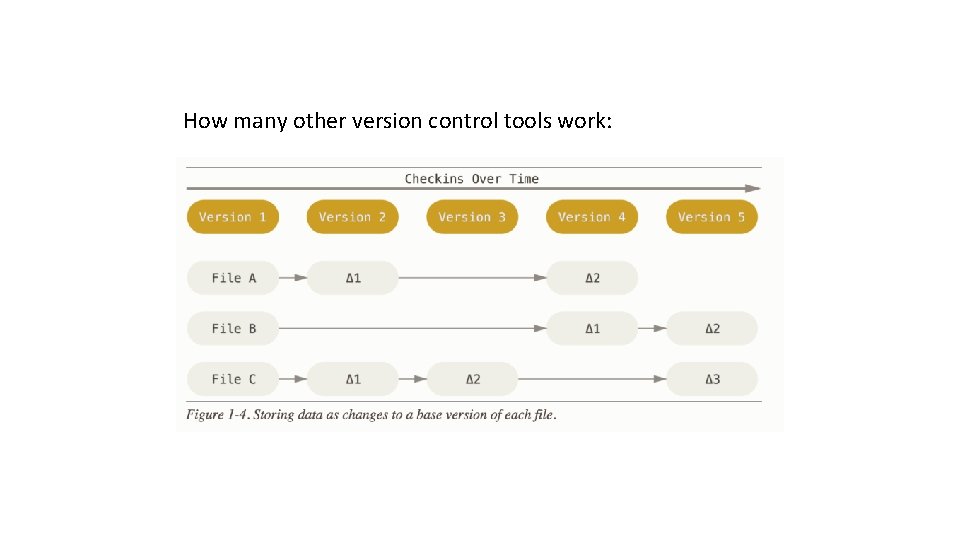
How many other version control tools work:

How Git works:

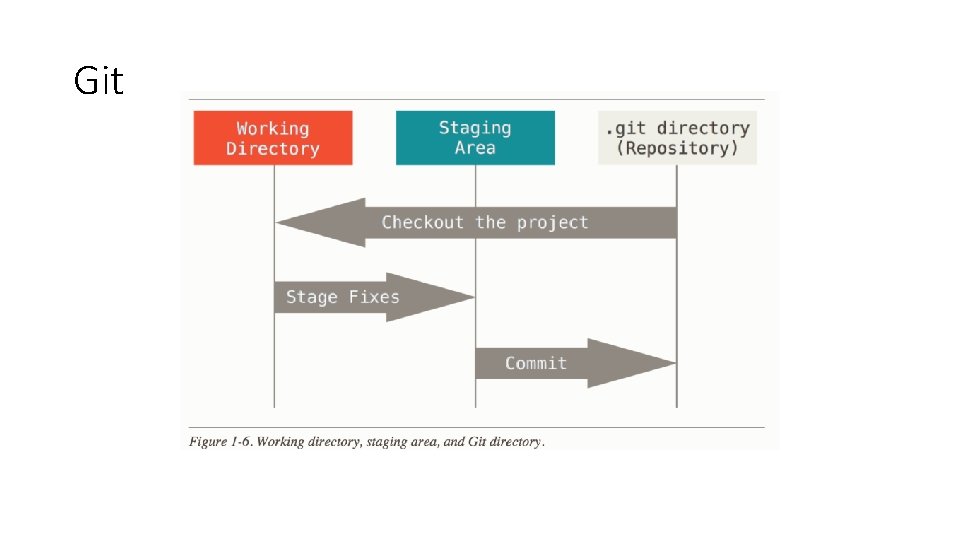
Git

Basic Git commands >git config >git help config >git init >git add *. c >git commit –m ‘initial project version’ >git status >git diff --staged >git commit -a -m 'added new benchmarks' >git clone https: //github. com/libgit 2 mylibgit
