UNIT I Overview of Transcription Translation and Recombinant
UNIT- I Overview of Transcription, Translation and Recombinant DNA Technology BY Anindya S. Ghosh Department of Biotechnology Indian Institute of Technology Kharagpur

Prokaryotic cells without a nucleus Eukaryotic cells with a nucleus • • • Nucleus Mitochondria Chloroplast Ribosomes RER SER Golgi body Cytoplasm Vacuoles • • • Cytoplasm Ribosomes Nuclear Zone DNA Plasmid Cell Membrane Mesosome Cell Wall Capsule (or slime layer) Flagellum
Macromolecules Protein Nucleic acids Olygosaccharides Lipids Complex macromolecules
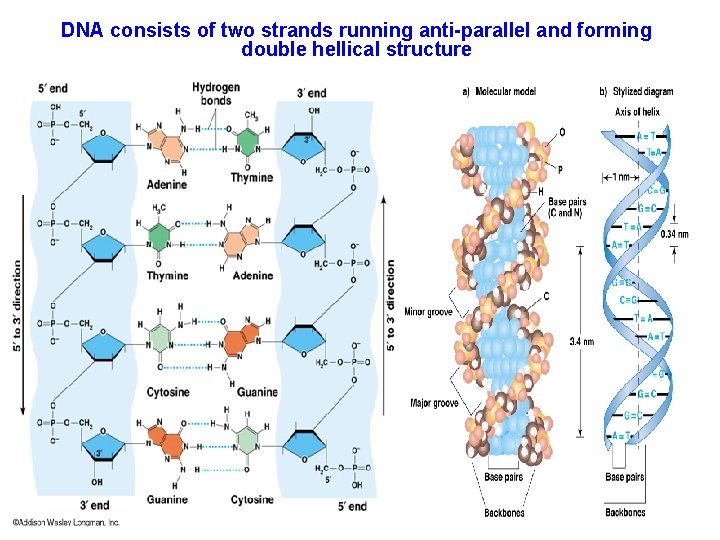
DNA consists of two strands running anti-parallel and forming double hellical structure
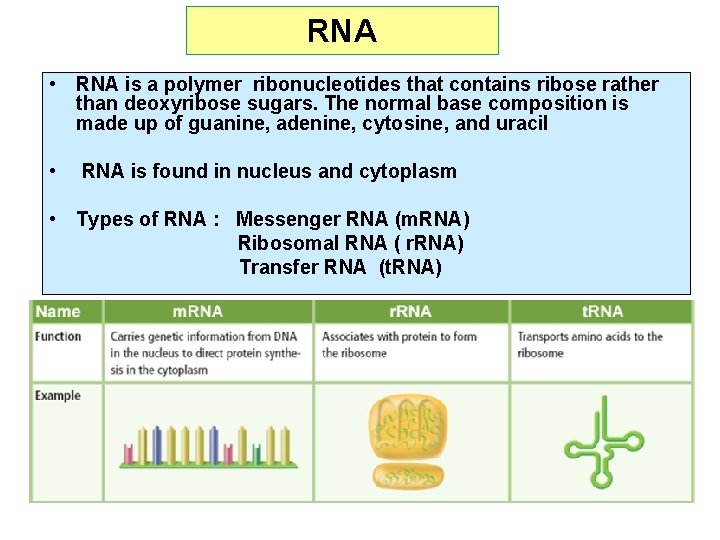
RNA • RNA is a polymer ribonucleotides that contains ribose rather than deoxyribose sugars. The normal base composition is made up of guanine, adenine, cytosine, and uracil • RNA is found in nucleus and cytoplasm • Types of RNA : Messenger RNA (m. RNA) Ribosomal RNA ( r. RNA) Transfer RNA (t. RNA)
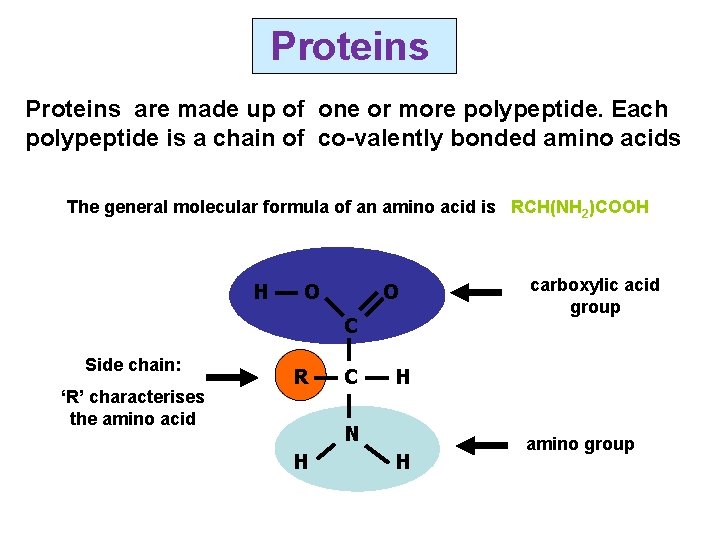
Proteins are made up of one or more polypeptide. Each polypeptide is a chain of co-valently bonded amino acids The general molecular formula of an amino acid is RCH(NH 2)COOH H O O C Side chain: ‘R’ characterises the amino acid R C H N H carboxylic acid group H amino group
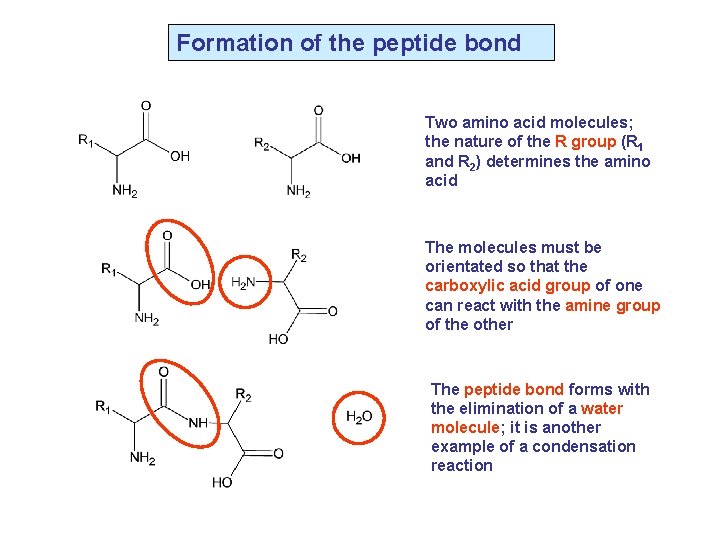
Formation of the peptide bond Two amino acid molecules; the nature of the R group (R 1 and R 2) determines the amino acid The molecules must be orientated so that the carboxylic acid group of one can react with the amine group of the other The peptide bond forms with the elimination of a water molecule; it is another example of a condensation reaction
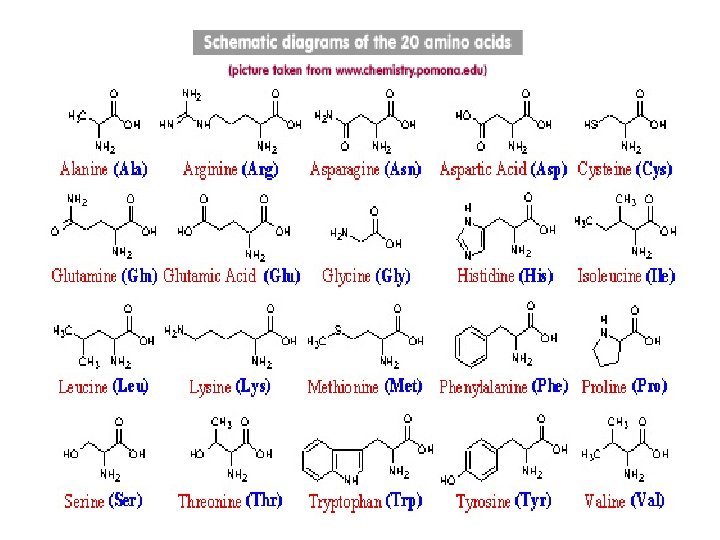

Single letter amino acid code G A Glycine Alanine Gly Ala P V Proline Valine Pro Val L Leucine Leu I Isoleucine Ile Met C Cysteine Cys F Phenylalanine Phe Y Tyrosine Tyr W Tryptophan Trp H Histidine His K Lysine Lys R Arginine Arg Gln N Asparagine Asn E Glutamic Acid Glu D Aspartic Acid Asp S Serine T Threonine Thr M Q Methionine Glutamine Ser
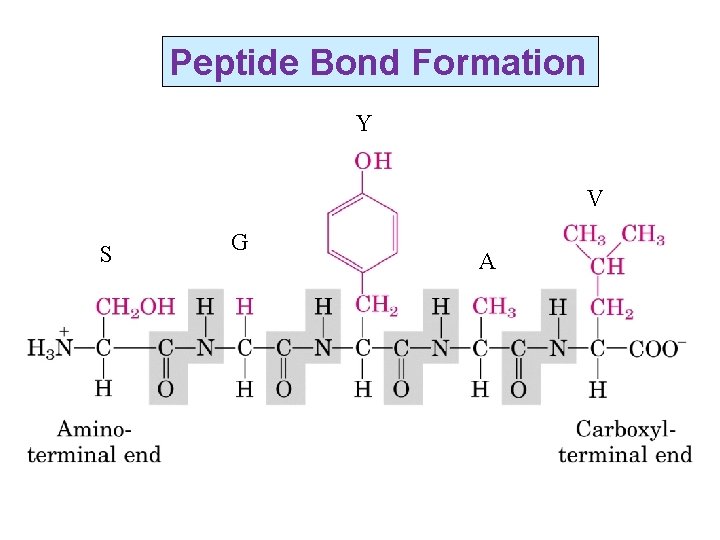
Peptide Bond Formation Y V S G A
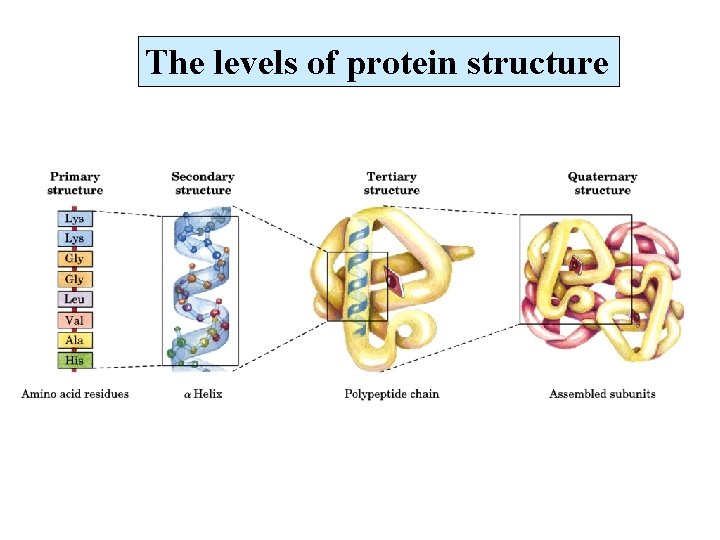
The levels of protein structure
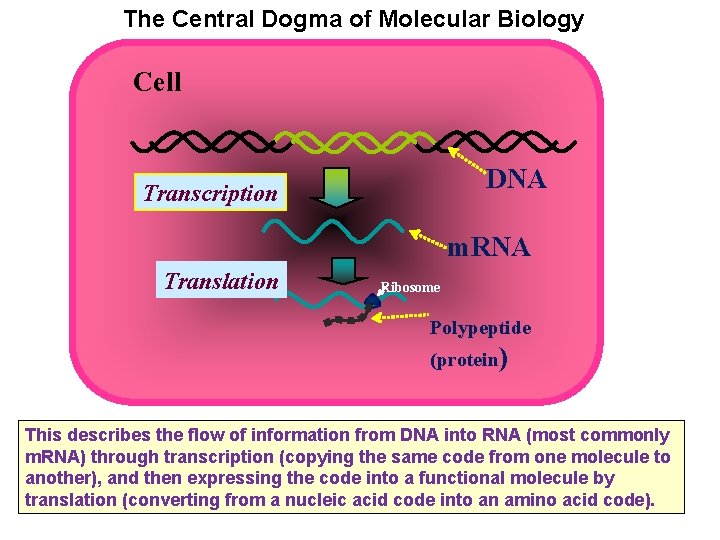
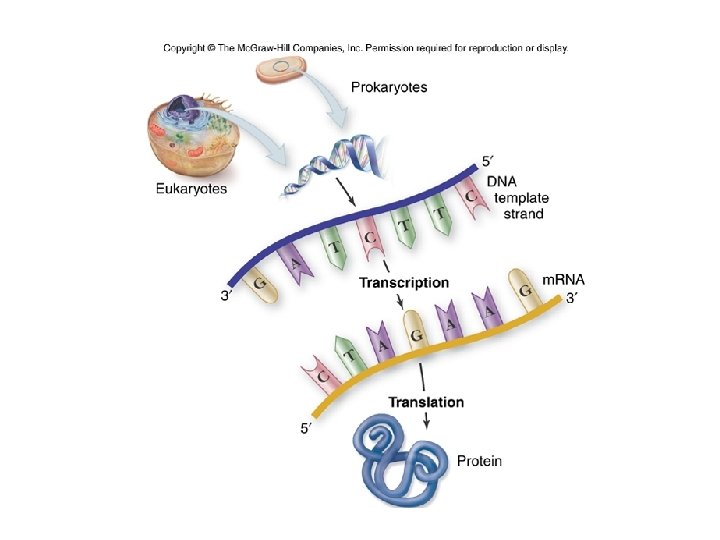
The Central Dogma of Molecular Biology Cell DNA Transcription m. RNA Translation Ribosome Polypeptide (protein) This describes the flow of information from DNA into RNA (most commonly m. RNA) through transcription (copying the same code from one molecule to another), and then expressing the code into a functional molecule by translation (converting from a nucleic acid code into an amino acid code).
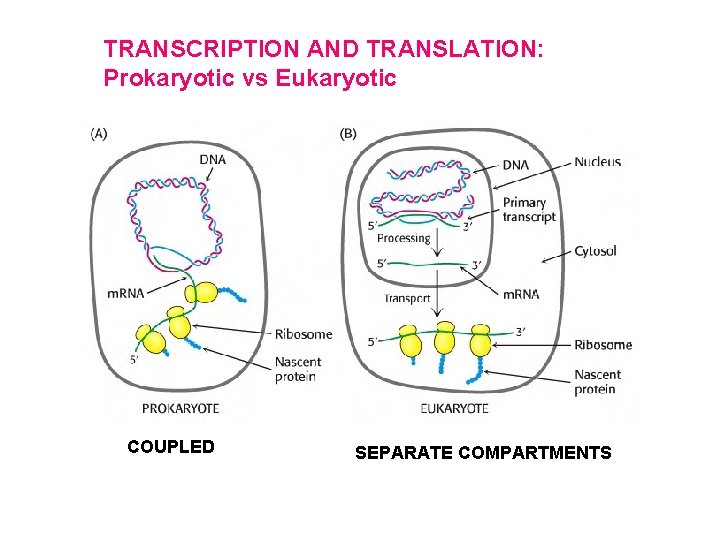
TRANSCRIPTION AND TRANSLATION: Prokaryotic vs Eukaryotic COUPLED SEPARATE COMPARTMENTS
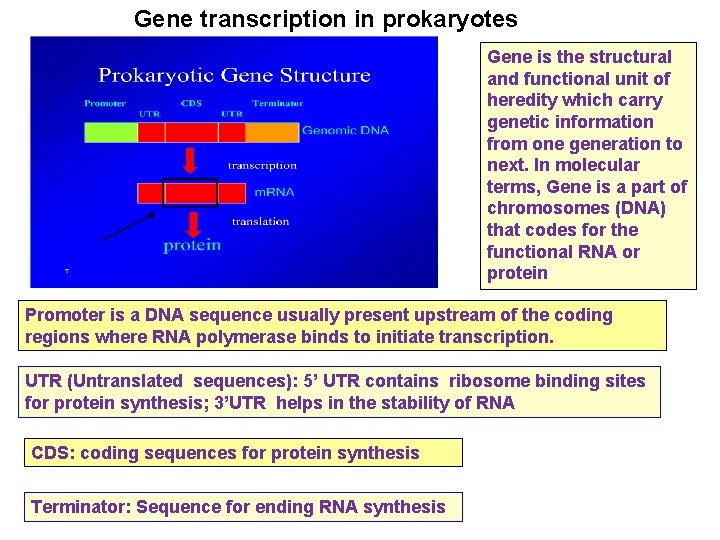
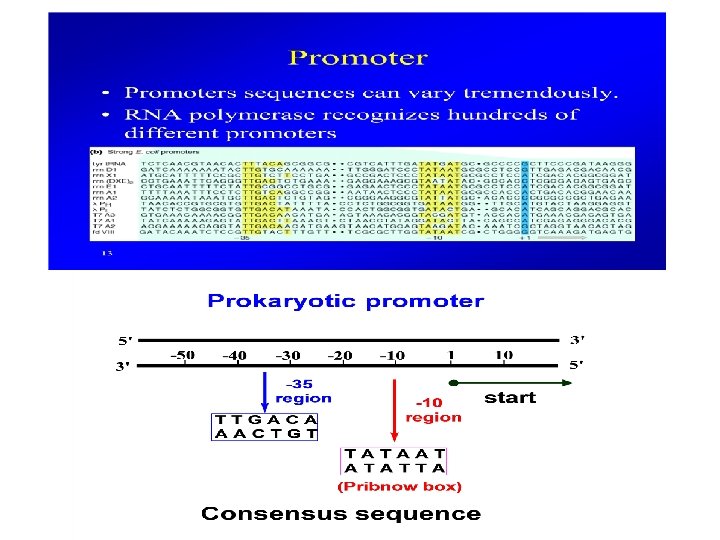
Gene transcription in prokaryotes Gene is the structural and functional unit of heredity which carry genetic information from one generation to next. In molecular terms, Gene is a part of chromosomes (DNA) that codes for the functional RNA or protein Promoter is a DNA sequence usually present upstream of the coding regions where RNA polymerase binds to initiate transcription. UTR (Untranslated sequences): 5’ UTR contains ribosome binding sites for protein synthesis; 3’UTR helps in the stability of RNA CDS: coding sequences for protein synthesis Terminator: Sequence for ending RNA synthesis
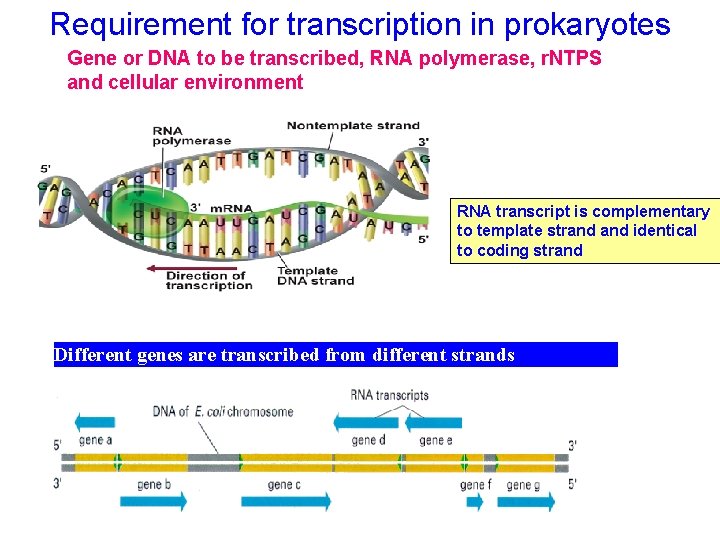
Requirement for transcription in prokaryotes Gene or DNA to be transcribed, RNA polymerase, r. NTPS and cellular environment RNA transcript is complementary to template strand identical to coding strand Different genes are transcribed from different strands
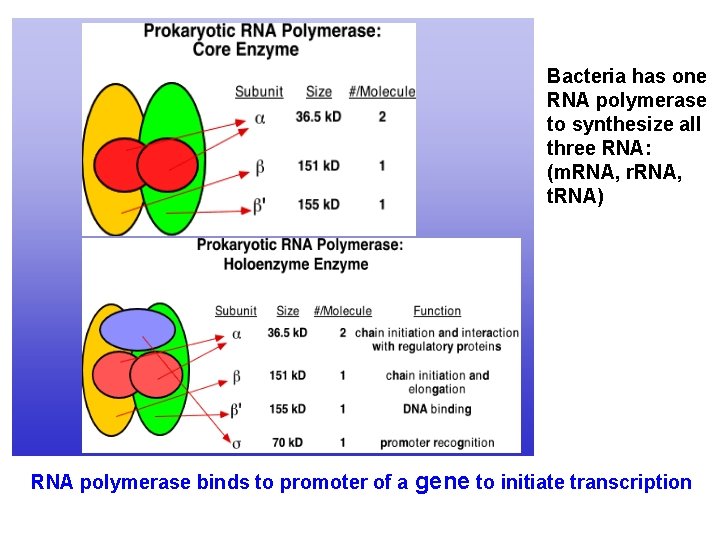
Bacteria has one RNA polymerase to synthesize all three RNA: (m. RNA, r. RNA, t. RNA) RNA polymerase binds to promoter of a gene to initiate transcription
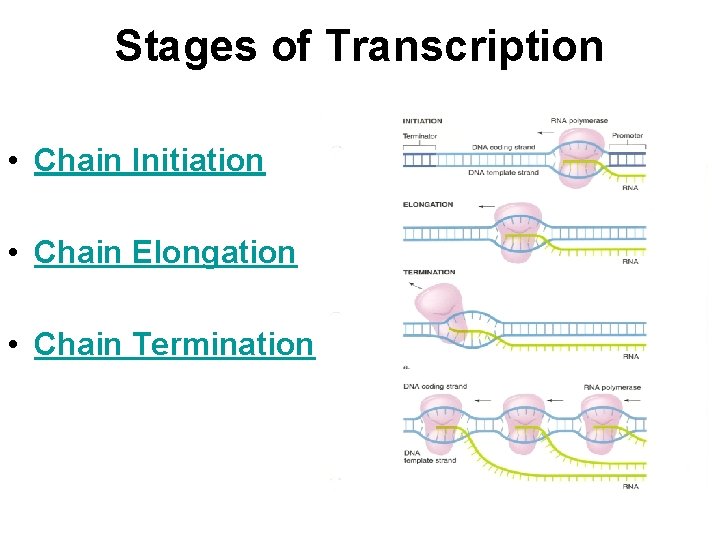
Stages of Transcription • Chain Initiation • Chain Elongation • Chain Termination
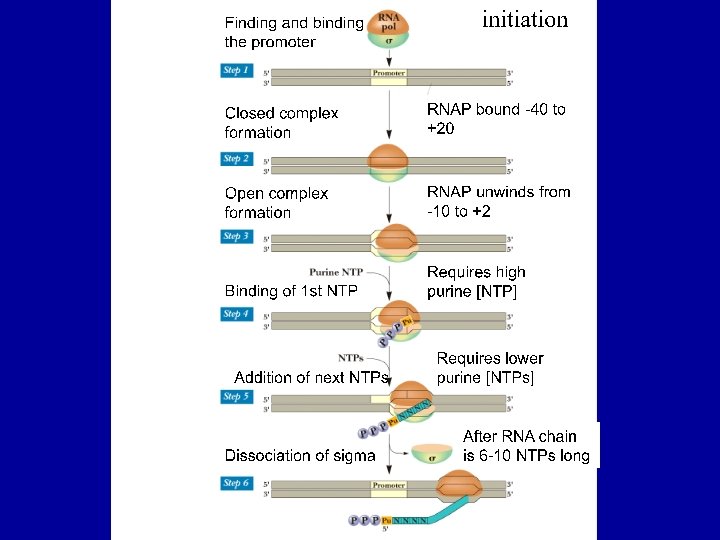
Events of Bacterial Transcription Initiation • Promoter recognition by RNA polymerase • Formation of Transcription Bubble by separating DNA strands • Bond formation between r. NTPs to start RNA synthesis • Escape of transcription apparatus from promoter
How RNA Polymerase finds promoter and Initiates Transcription • Core enzyme can synthesize RNA on a DNA template but cannot initiate transcription. • Core polymerase loosely binds at random sites in DNA without discriminating promoter and other sequences. • Binding of sigma in the polymerase (holoenzyme) reduced the chance of binding loosely, and the enzymes moves along the DNA. • When it reaches the promoter sequences, sigma factor recognize specifically -35 sequence and binds tightly (-40 to +20 regions). Then it unwinds DNA (17 bp) from -10 regions and adds ribonucleotide (G or A) in the +1 site. • After the synthesis of 6 -9 nucleotides long RNA without movement of enzyme [abortive initiation], sigma factor falls off from holoenzyme and the core enzyme enters the elongation process [promoter escape]
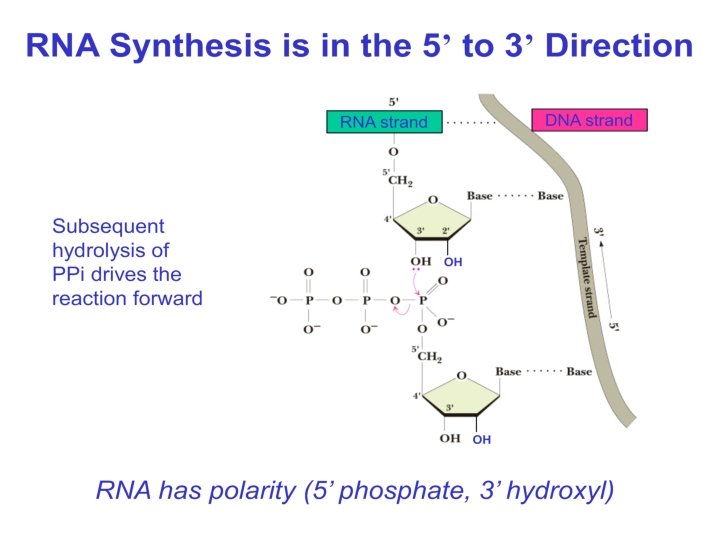
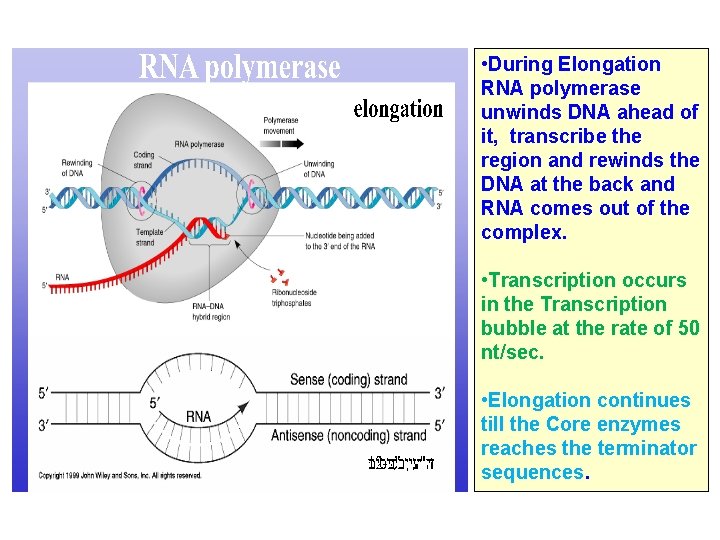
• During Elongation RNA polymerase unwinds DNA ahead of it, transcribe the region and rewinds the DNA at the back and RNA comes out of the complex. • Transcription occurs in the Transcription bubble at the rate of 50 nt/sec. • Elongation continues till the Core enzymes reaches the terminator sequences.
Transcription Termination Transcription ends after a terminator is transcribed • Two types of terminators in bacteria: – Rho-dependent terminators – Rho-independent terminators
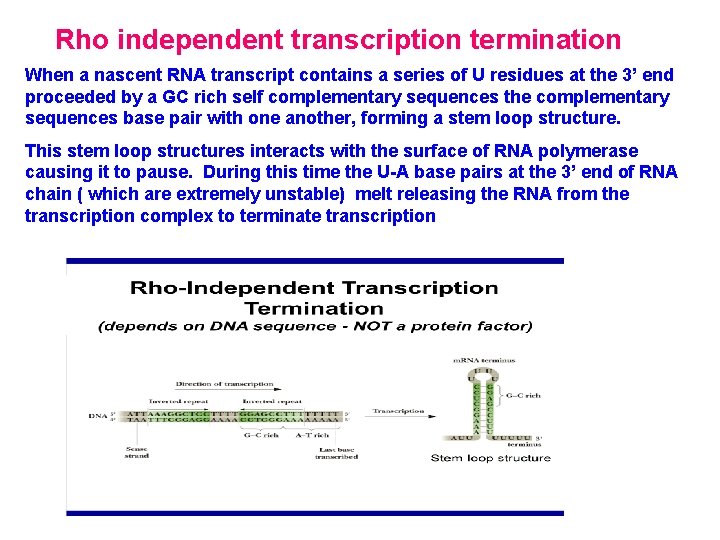
Rho independent transcription termination When a nascent RNA transcript contains a series of U residues at the 3’ end proceeded by a GC rich self complementary sequences the complementary sequences base pair with one another, forming a stem loop structure. This stem loop structures interacts with the surface of RNA polymerase causing it to pause. During this time the U-A base pairs at the 3’ end of RNA chain ( which are extremely unstable) melt releasing the RNA from the transcription complex to terminate transcription
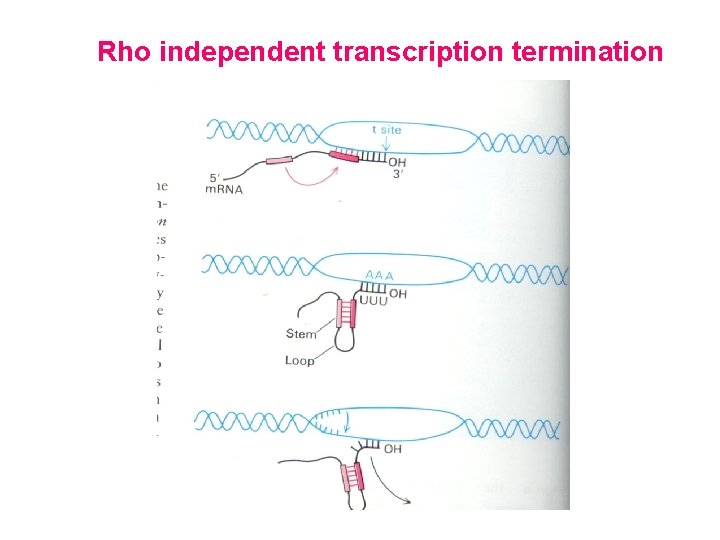
Rho independent transcription termination
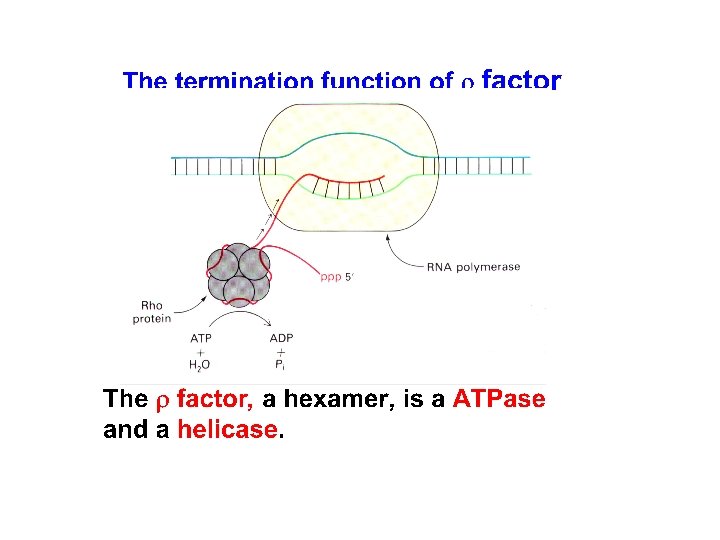
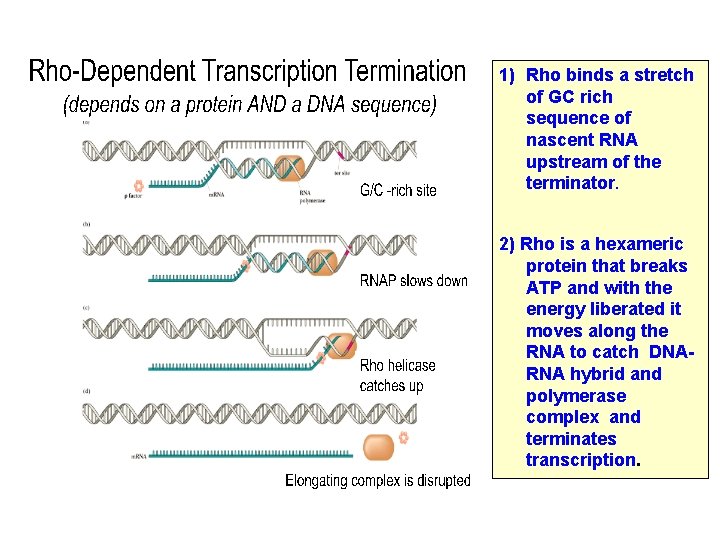
1) Rho binds a stretch of GC rich sequence of nascent RNA upstream of the terminator. 2) Rho is a hexameric protein that breaks ATP and with the energy liberated it moves along the RNA to catch DNARNA hybrid and polymerase complex and terminates transcription.
Translation: Protein synthesis Translation is the process of decoding a m. RNA molecule into a polypeptide chain or protein
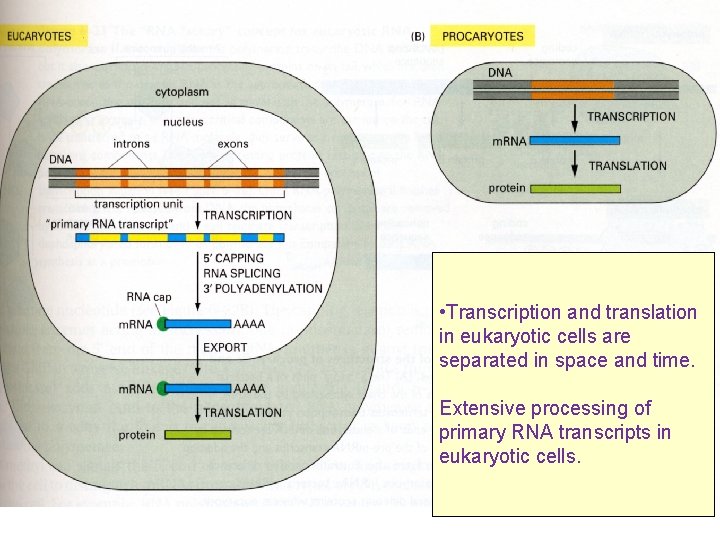
• Transcription and translation in eukaryotic cells are separated in space and time. Extensive processing of primary RNA transcripts in eukaryotic cells.
Translation It is process of protein synthesis (assembly of amino acids) using m. RNA as template with the help of t. RNA and ribosomes (r. RNA with several protein). Therefore it requires the participation of multiple types of RNA: • messenger RNA (m. RNA) carries the information from DNA that encodes proteins • ribosomal RNA (r. RNA) is a structural component of the ribosome • transfer RNA (t. RNA) carries amino acids to the ribosome for translation
The Genetic Code The genetic code is the way in which the nucleotide sequence in nucleic acids specifies the amino acid sequence in proteins. A codon is a set of 3 nucleotides that specifies a particular amino acid. Therefore, m. RNA carries information from DNA in a three letter genetic code. • A three-letter code is used because there are 20 different amino acids that are used to make proteins. • If a two-letter code were used there would not be enough codons to select all 20 amino acids. • That is, there are 4 bases in RNA, so 42 (4 x 4)=16; where as for 3 lettered code the number is 43 (4 x 4 x 4)=64.
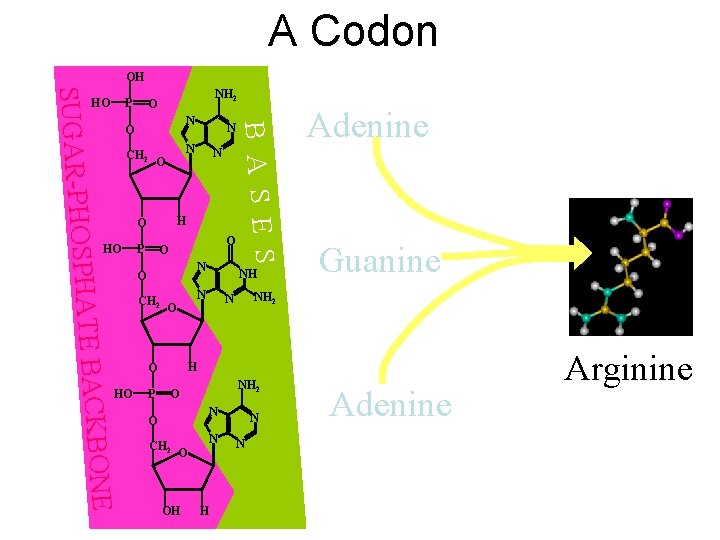
A Codon OH NH 2 O N O CH 2 N O P O O N CH 2 P NH N O Adenine Guanine NH 2 N H O ONE HATE BACKB O HO N H O HO N B A S E S SUGAR-PHOSP P HO NH 2 O N O CH 2 O OH N N Adenine Arginine
GENETIC CODE Therefore, there is a total of 64 codons with m. RNA, 61 specify particular amino acid. The remaining three codons (UAA, UAG, & UGA) are stop codons, which signify the end of a polypeptide chain (protein). This means there are more than one codon for each of the 20 amino acids. Besides selecting the amino acid methionine, the codon AUG also serves as the “initiator” codon, which starts the synthesis of a protein
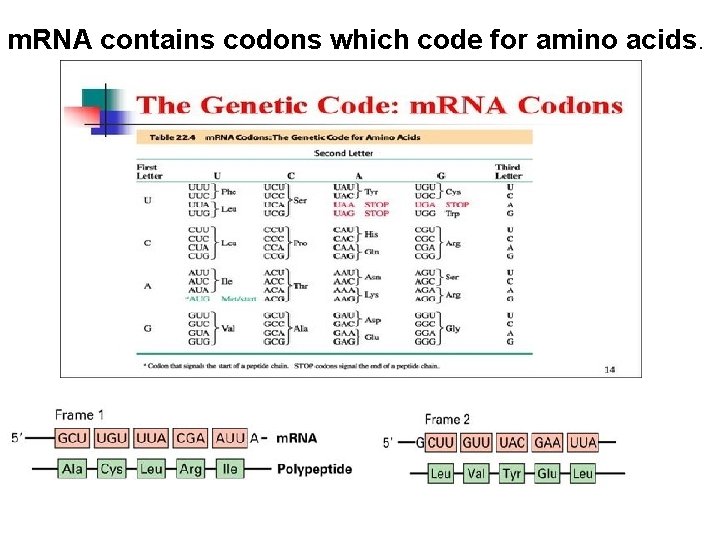
m. RNA contains codons which code for amino acids.
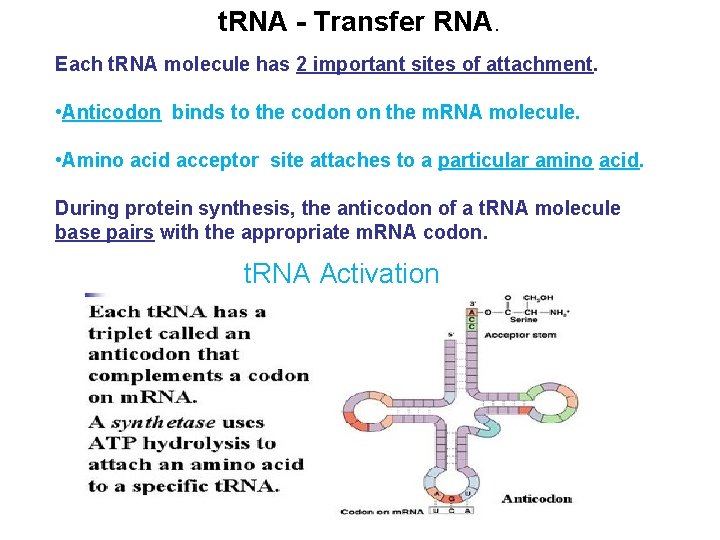
t. RNA - Transfer RNA. Each t. RNA molecule has 2 important sites of attachment. • Anticodon binds to the codon on the m. RNA molecule. • Amino acid acceptor site attaches to a particular amino acid. During protein synthesis, the anticodon of a t. RNA molecule base pairs with the appropriate m. RNA codon. t. RNA Activation
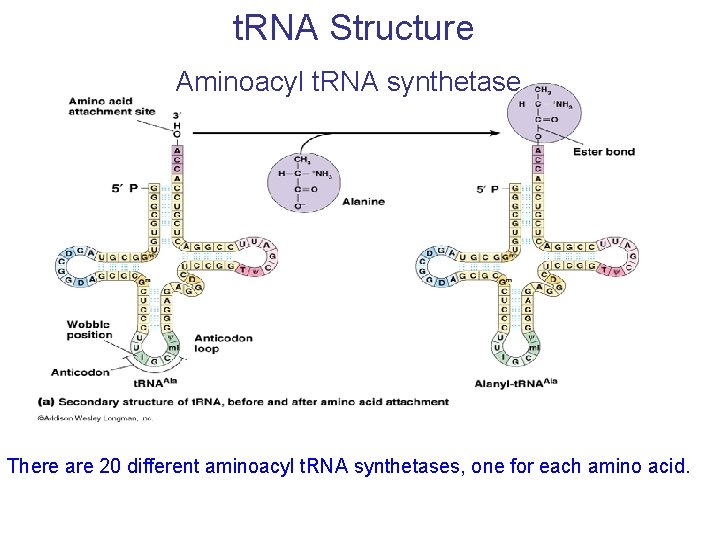
t. RNA Structure Aminoacyl t. RNA synthetase There are 20 different aminoacyl t. RNA synthetases, one for each amino acid.
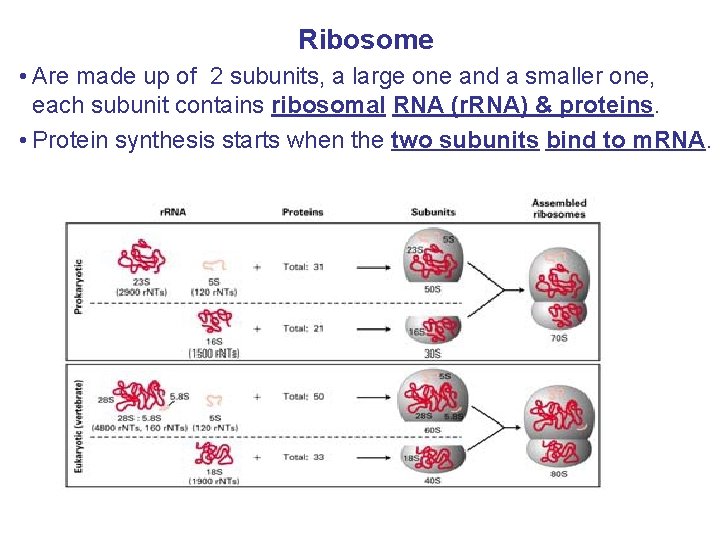
Ribosome • Are made up of 2 subunits, a large one and a smaller one, each subunit contains ribosomal RNA (r. RNA) & proteins. • Protein synthesis starts when the two subunits bind to m. RNA.
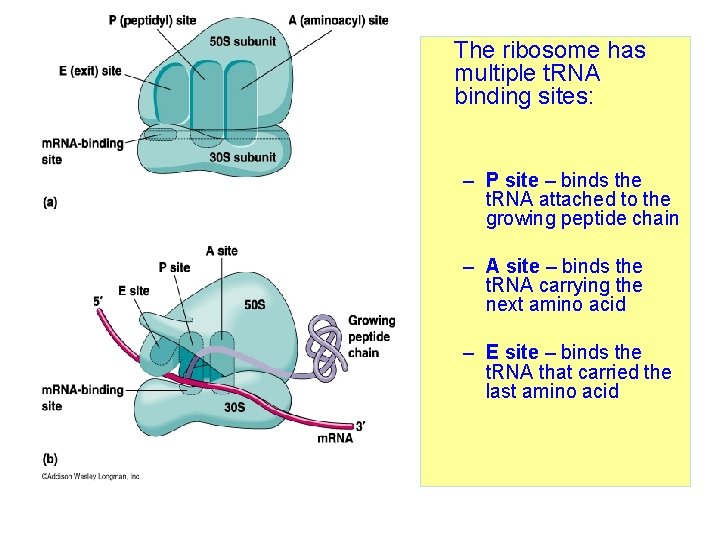
The ribosome has multiple t. RNA binding sites: – P site – binds the t. RNA attached to the growing peptide chain – A site – binds the t. RNA carrying the next amino acid – E site – binds the t. RNA that carried the last amino acid
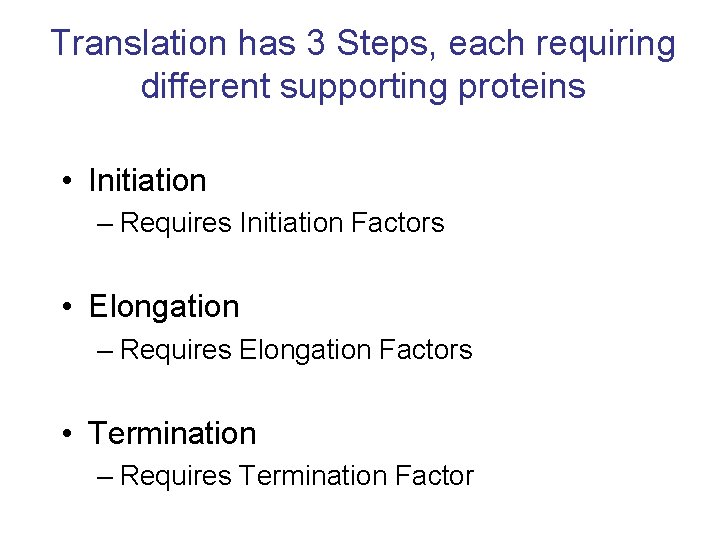
Translation has 3 Steps, each requiring different supporting proteins • Initiation – Requires Initiation Factors • Elongation – Requires Elongation Factors • Termination – Requires Termination Factor
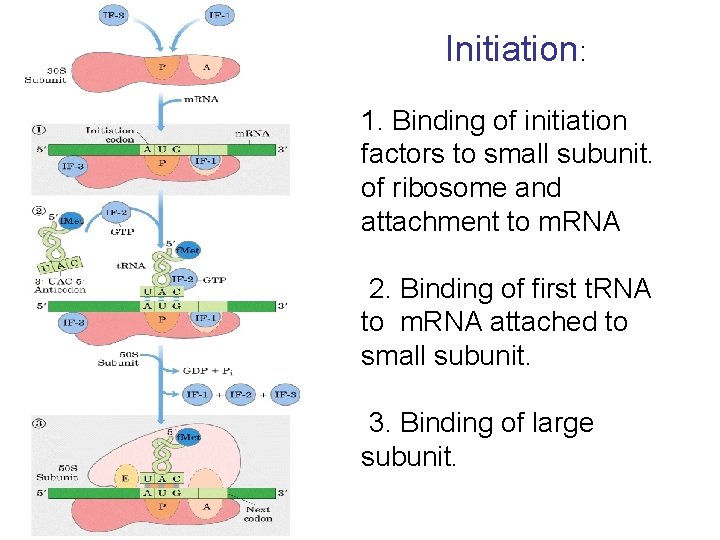
Initiation: 1. Binding of initiation factors to small subunit. of ribosome and attachment to m. RNA 2. Binding of first t. RNA to m. RNA attached to small subunit. 3. Binding of large subunit.
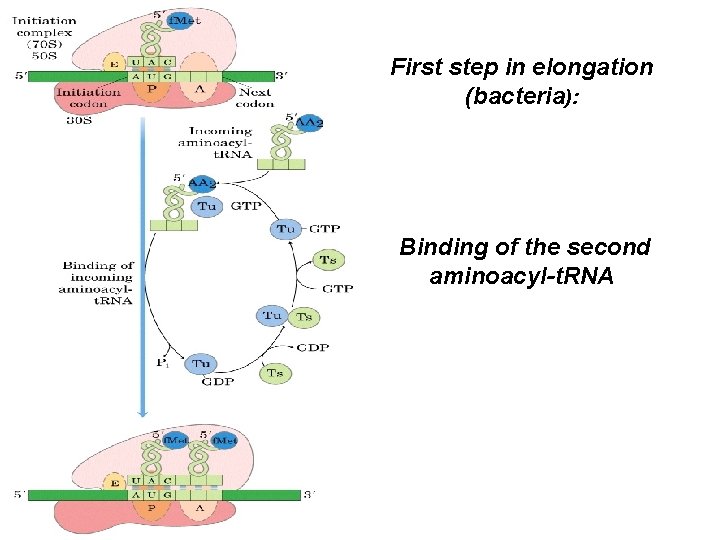
First step in elongation (bacteria): Binding of the second aminoacyl-t. RNA
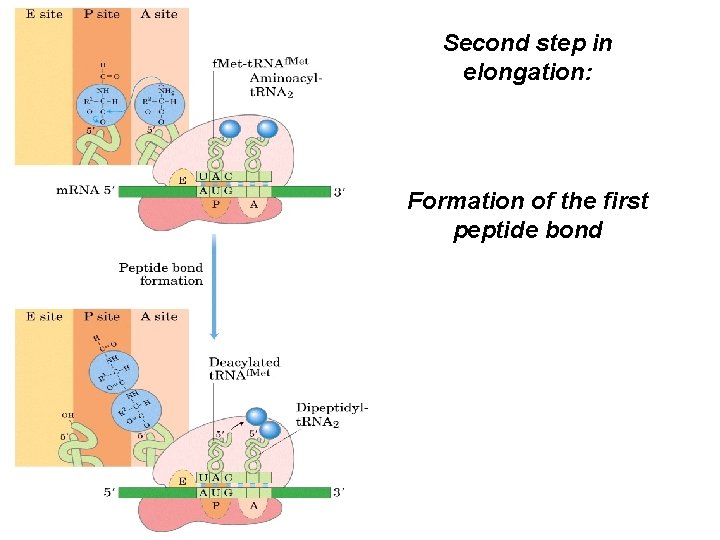
Second step in elongation: Formation of the first peptide bond
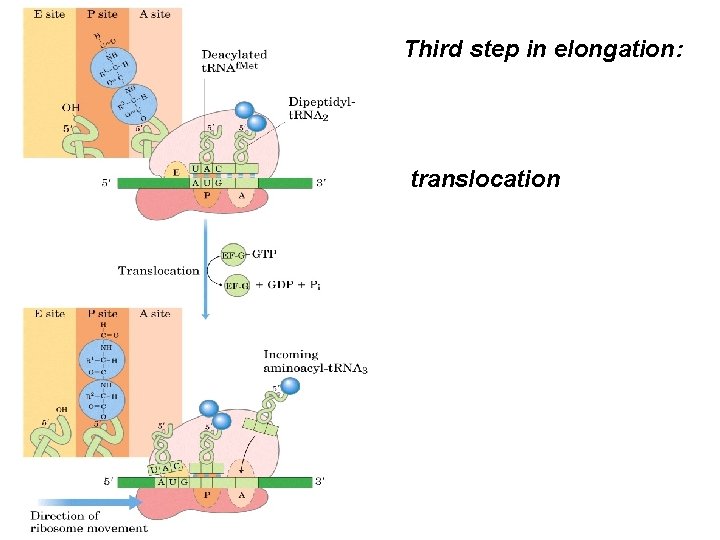
Third step in elongation: translocation
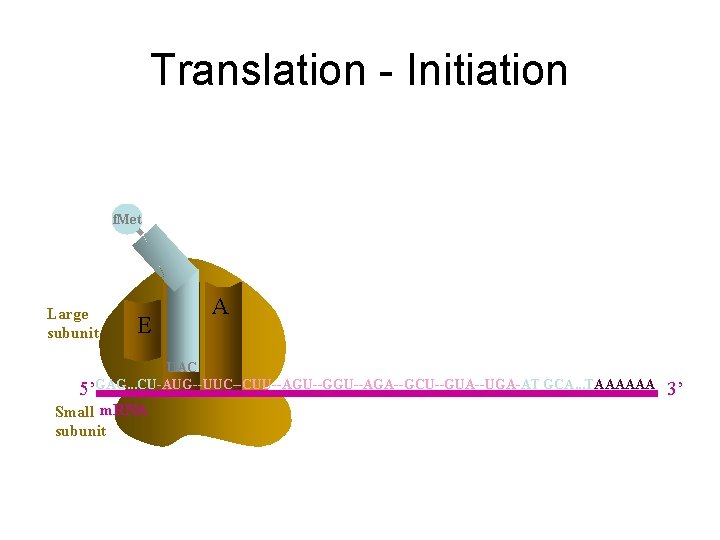
Translation - Initiation f. Met Large subunit E P A UAC 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA Small m. RNA subunit 3’
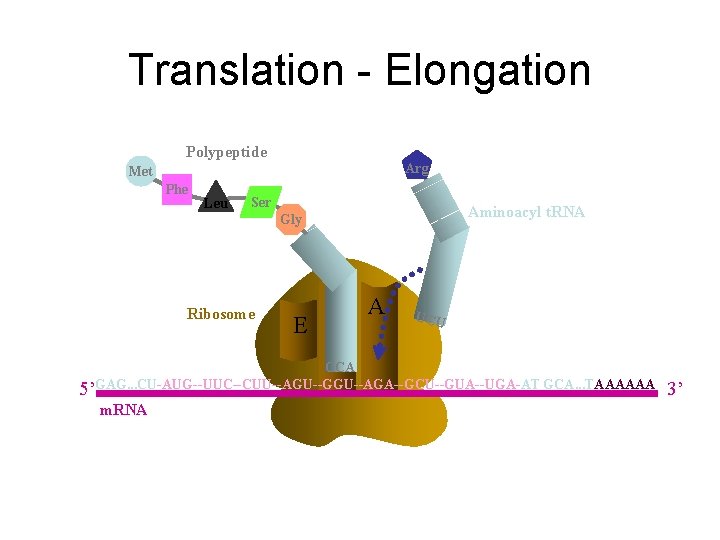
Translation - Elongation Polypeptide Arg Met Phe Leu Ser Aminoacyl t. RNA Gly Ribosome E P A UCU CCA 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’
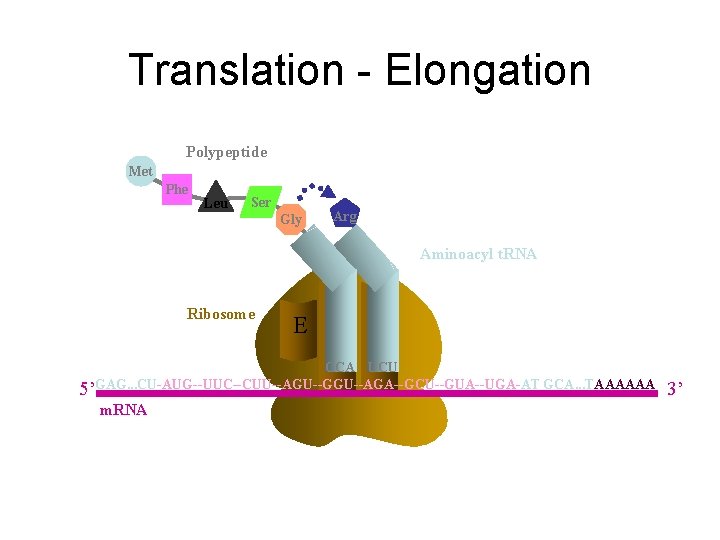
Translation - Elongation Polypeptide Met Phe Leu Ser Gly Arg Aminoacyl t. RNA Ribosome E P A CCA UCU 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’
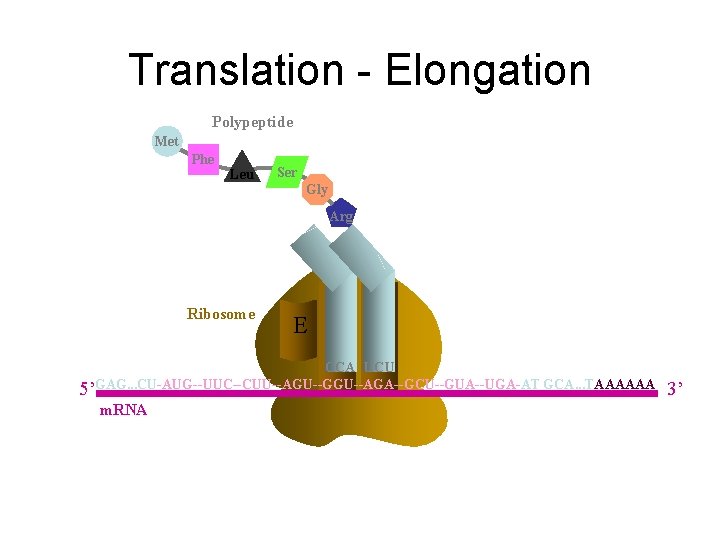
Translation - Elongation Polypeptide Met Phe Leu Ser Gly Arg Ribosome E P A CCA UCU 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’
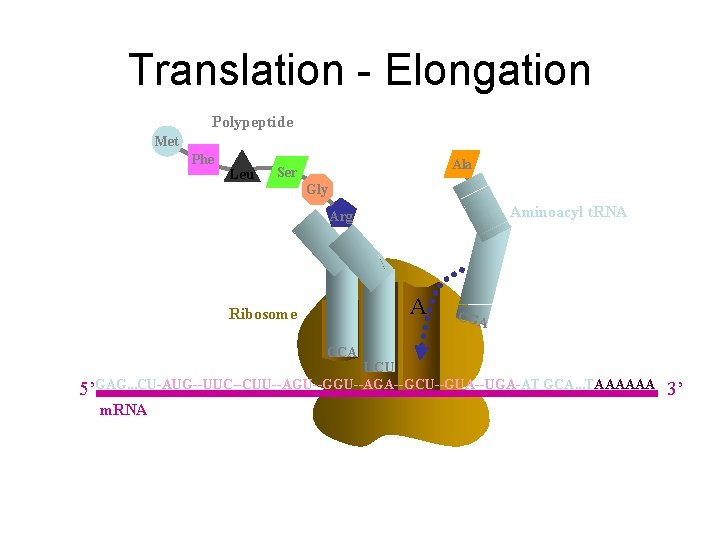
Translation - Elongation Polypeptide Met Phe Leu Ala Ser Gly Aminoacyl t. RNA Arg Ribosome E P A CGA CCA UCU 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’
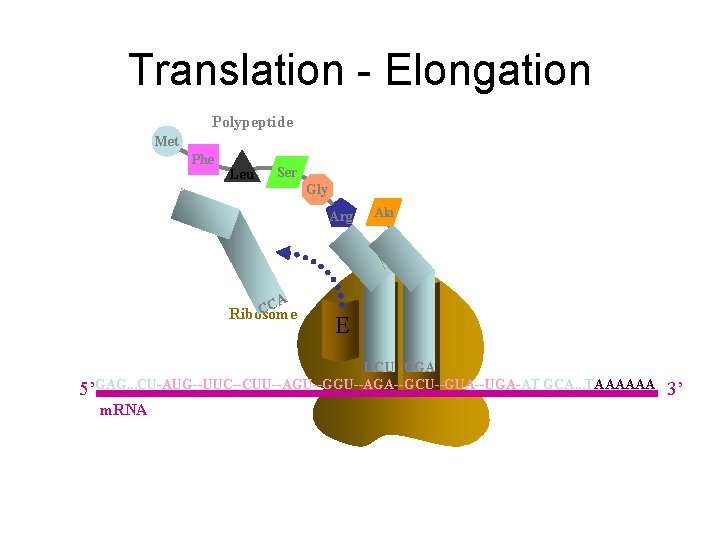
Translation - Elongation Polypeptide Met Phe Leu Ser Gly Arg CA C Ribosome E Ala P A UCU CGA 5’GAG. . . CU-AUG--UUC--CUU--AGU--GGU--AGA--GCU--GUA--UGA-AT GCA. . . TAAAAAA m. RNA 3’
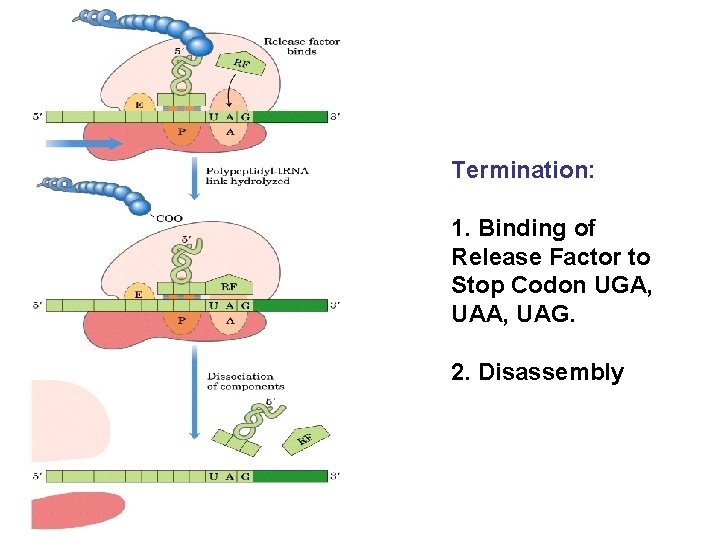
Termination: 1. Binding of Release Factor to Stop Codon UGA, UAG. 2. Disassembly
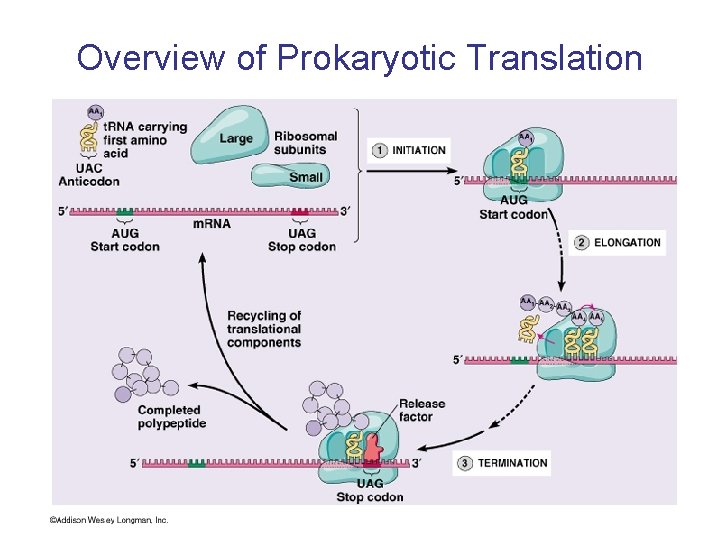
Overview of Prokaryotic Translation
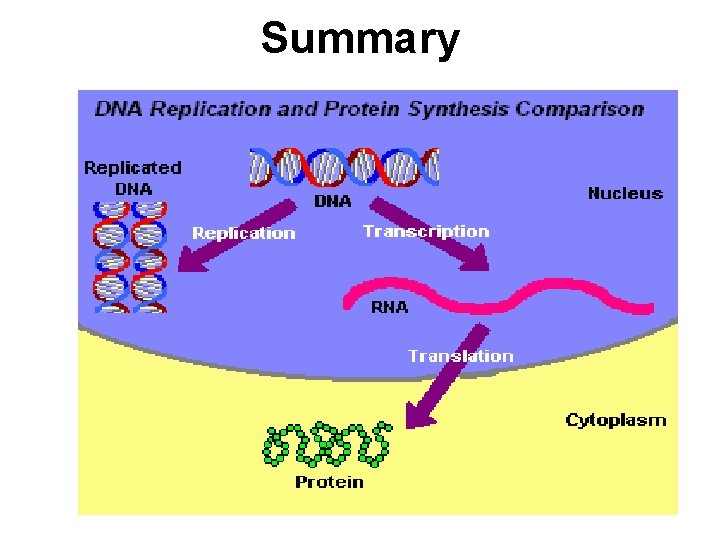
Summary
Recombinant DNA • Production of a unique DNA molecule by joining together two or more DNA fragments not normally associated with each other • DNA fragments are usually derived from different biological sources • A series of procedures used to recombine DNA segments and are called Recombinant DNA Technology
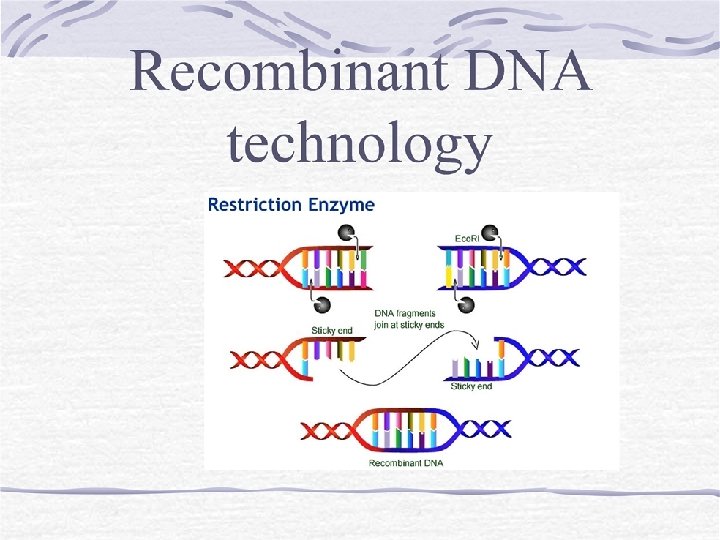
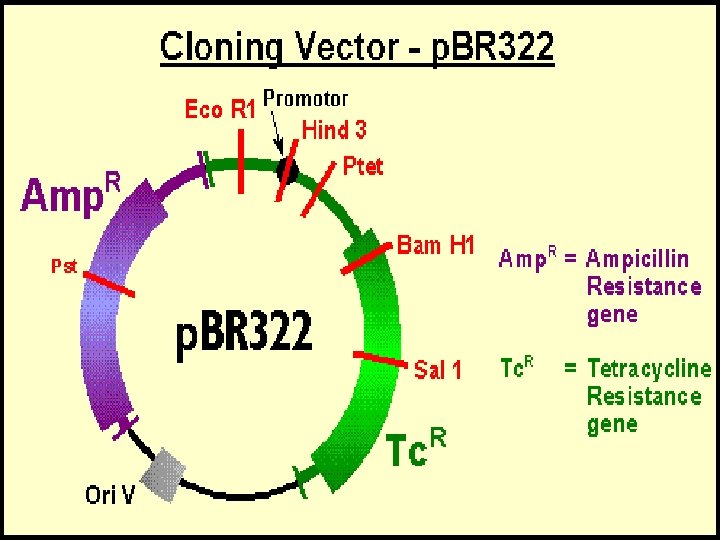
Basic principle of recombinant DNA technology One DNA molecule (called insert) is isolated from one sources and then this DNA is inserted into another DNA molecule called ‘vector’ Mostly bacterial plasmid is used as vector The recombinant vector is then introduced into a host cell where it replicates, transcribes and translates to produce protein.

Basic steps in. Recobinant DNA Technology 1. Isolate the gene 2. Insert it in a host using a vector (plasmid) 3. Produce as many copies of the host as possible 4. Separate and purify the product of the gene
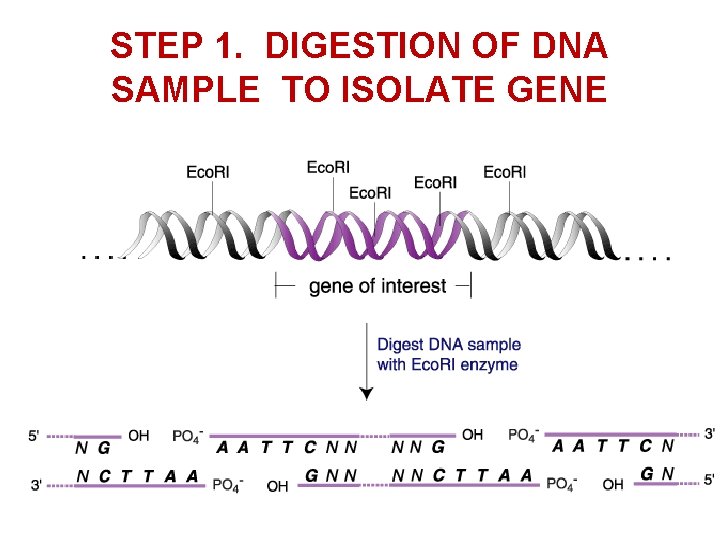
STEP 1. DIGESTION OF DNA SAMPLE TO ISOLATE GENE
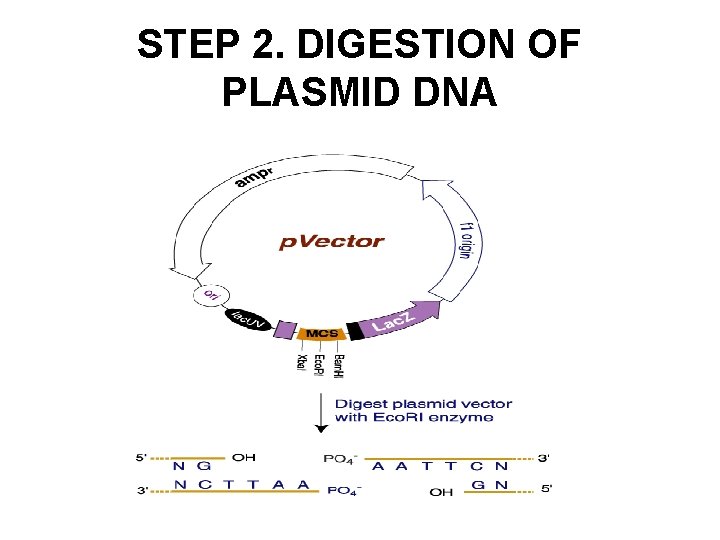
STEP 2. DIGESTION OF PLASMID DNA
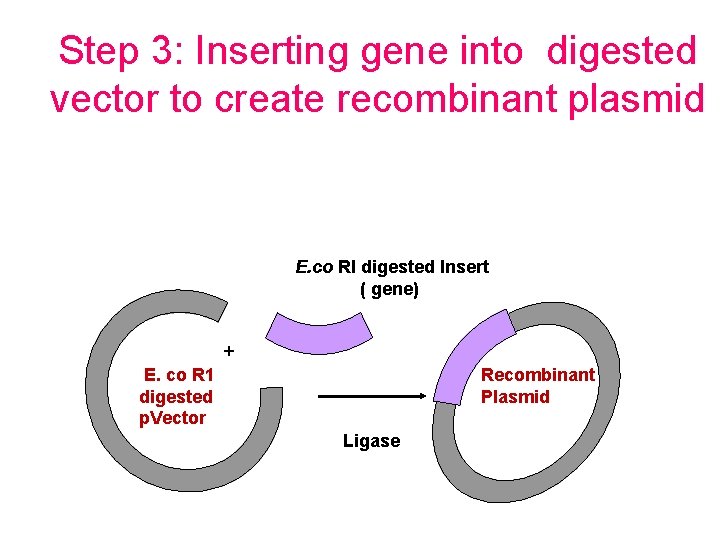
Step 3: Inserting gene into digested vector to create recombinant plasmid E. co RI digested Insert ( gene) + E. co R 1 digested p. Vector Recombinant Plasmid Ligase
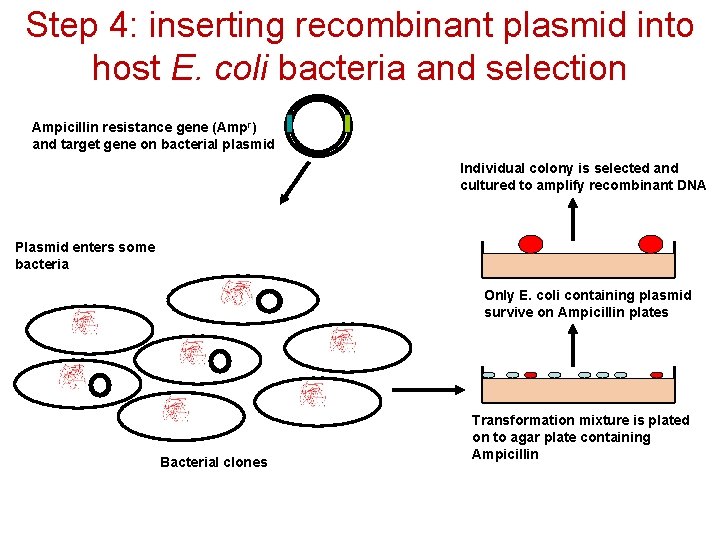
Step 4: inserting recombinant plasmid into host E. coli bacteria and selection Ampicillin resistance gene (Ampr) and target gene on bacterial plasmid Individual colony is selected and cultured to amplify recombinant DNA Plasmid enters some bacteria Only E. coli containing plasmid survive on Ampicillin plates Bacterial clones Transformation mixture is plated on to agar plate containing Ampicillin
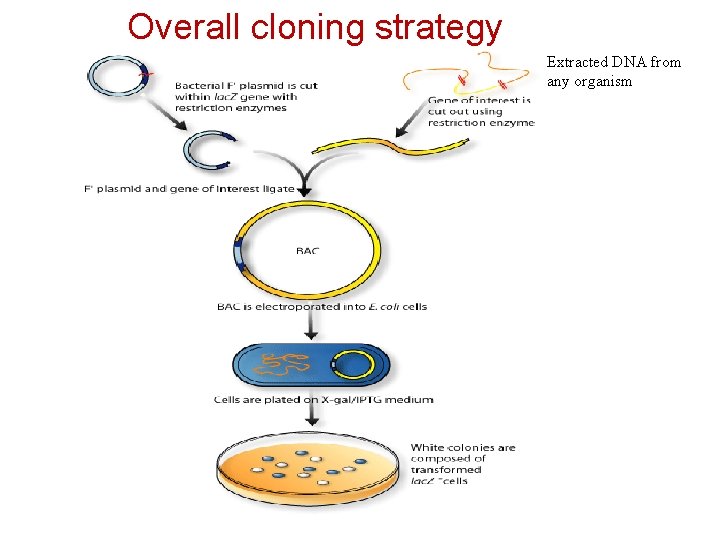
Overall cloning strategy Extracted DNA from any organism
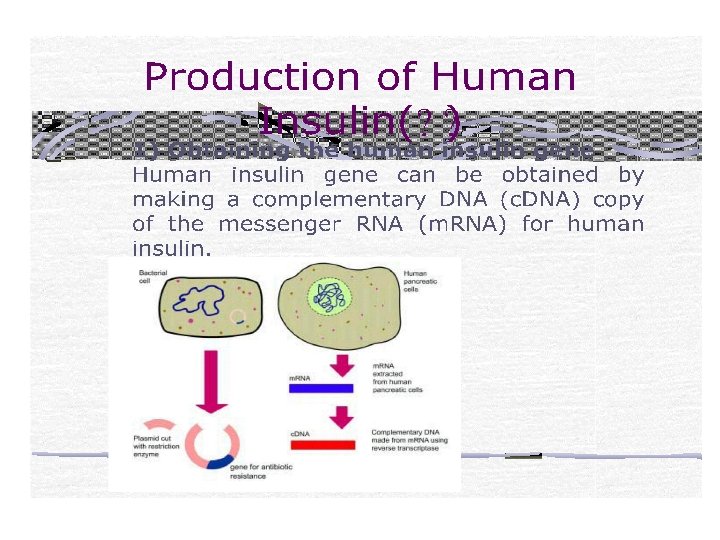



- Slides: 70