Uncorrelated and Autocorrelated relaxed phylogenetics Michal DefoinPlatel and





















- Slides: 35

Uncorrelated and Autocorrelated relaxed phylogenetics Michaël Defoin-Platel and Alexei Drummond Juin 2008 bioinf. cs. auckland. ac. nz

(Bayesian) RELAXED PHYLOGENETICS Relaxed Phylogenetics allows • the co-estimation of divergence times together with a phylogenetic reconstruction • should be compared with t 0 b 2 b 5 b 3 t 2 time b 1 t 1 b 4 Unrooted (2 n-3 parameters) Rooted with a strict clock (n-1 divergence times) Relaxed Phylogenetics 2

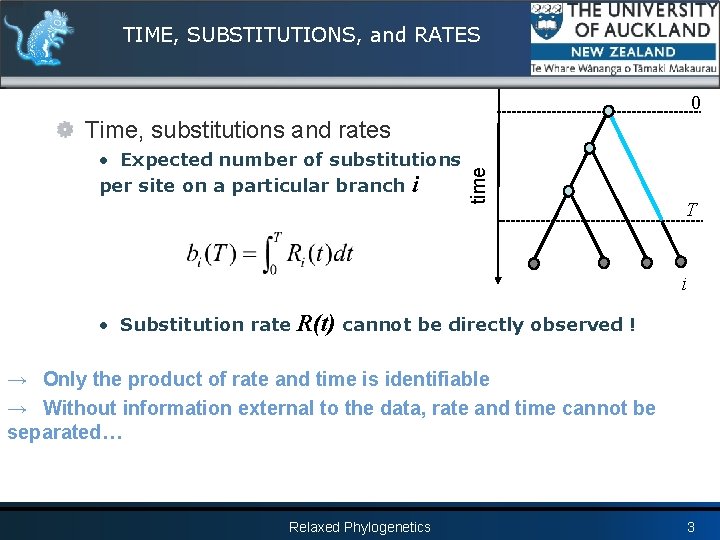
TIME, SUBSTITUTIONS, and RATES 0 • Expected number of substitutions per site on a particular branch i time Time, substitutions and rates T i • Substitution rate R(t) cannot be directly observed ! → Only the product of rate and time is identifiable → Without information external to the data, rate and time cannot be separated… Relaxed Phylogenetics 3

MOLECULAR CLOCK HYPOTHESIS Molecular Clock Hypothesis (MCH) (Zuckerlandl and Pauling 1965) • DNA and protein sequences change at a rate that is constant over time • First the substitution rate is estimated then time corresponds to sequence divergence divided by the rate → Estimation of relative rate and relative divergence times Calibration • Time reference, scaling • Bayesian Phylogenetics : Priors on node height or on tips → Transform relative to absolute rate Relaxed Phylogenetics 4

MOLECULAR CLOCK HYPOTHESIS Substitution rate depends on • Natural selection, population size, body mass, generation time, mutation rate, mutation pattern, … → MCH is often violated ! How to deal with non-clock like data • Keep them ! • Remove them ! • Relax the MCH → Allow the rate of evolution to vary → Make assumptions about the variations Relaxed Phylogenetics 5

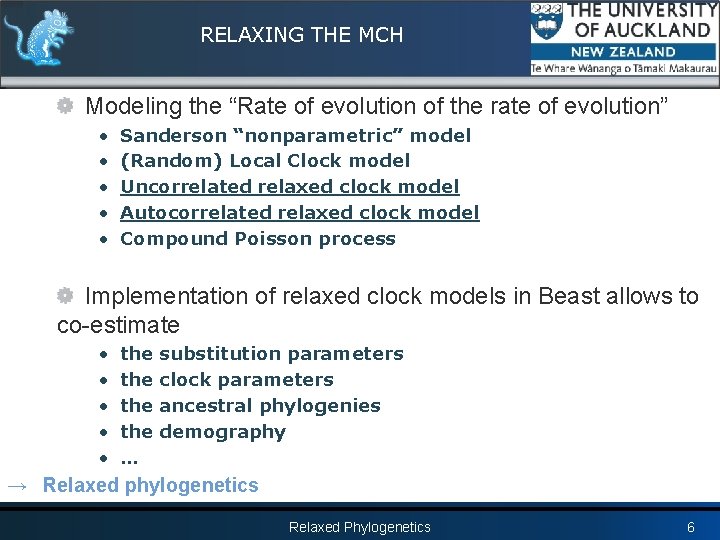
RELAXING THE MCH Modeling the “Rate of evolution of the rate of evolution” • • • Sanderson “nonparametric” model (Random) Local Clock model Uncorrelated relaxed clock model Autocorrelated relaxed clock model Compound Poisson process Implementation of relaxed clock models in Beast allows to co-estimate • • • the the … substitution parameters clock parameters ancestral phylogenies demography → Relaxed phylogenetics Relaxed Phylogenetics 6

UNCORRELATED RELAXED CLOCK (UC) Drummond et al 2006 Hypothesis • The rate of evolution is probably never exactly the same for all evolutionary lineages • Rates follow a given distribution Prior on rates → Distribution of the rates given by the hyperparameters and 2 or Relaxed Phylogenetics 7

UNCORRELATED RELAXED CLOCK (UC) Drummond et al 2006 Implementation r 0 • The distribution is discretized • Each branch of the tree is assigned a given rate category • Category mixing : swapped drawn (uniform) random walk 0 Relaxed Phylogenetics t 1 r 2 time • Different rates in a tree • But a constant rate per branch • On a given rooted tree of n species 2 n-2 rates n-1 divergence times t 0 r 1 r 4 r 5 1 2 2 4 t 2 r 3 3 6 relative rate r 4 8 10 8

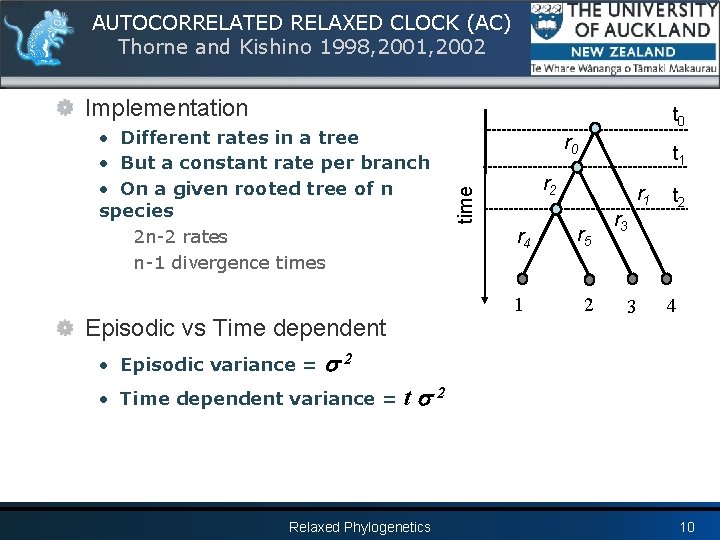
AUTOCORRELATED RELAXED CLOCK (AC) Thorne and Kishino 1998, 2001, 2002 Hypothesis • The rate is probably never exactly the same for all evolutionary lineages • For closely related lineages the rates should be similar r. A Prior on rates t r • log of the rates follow a Normal distribution • Expectation of a rate r is its ancestor rate r. A → Rate at the root node is given by the hyperparameter → Amount of variation is given by the hyperparameter 2 Relaxed Phylogenetics 9

AUTOCORRELATED RELAXED CLOCK (AC) Thorne and Kishino 1998, 2001, 2002 Implementation Episodic vs Time dependent • Episodic variance = r 0 t 1 r 2 time • Different rates in a tree • But a constant rate per branch • On a given rooted tree of n species 2 n-2 rates n-1 divergence times t 0 r 1 r 4 r 5 1 2 r 3 3 t 2 4 2 • Time dependent variance = t 2 Relaxed Phylogenetics 10

GOALS of this TALK Validation of models implementation Comparison of models • • • Fit the data Deal with calibrations Estimate of divergence times Estimate of rates Reconstruct the tree topology Relaxed Phylogenetics 11

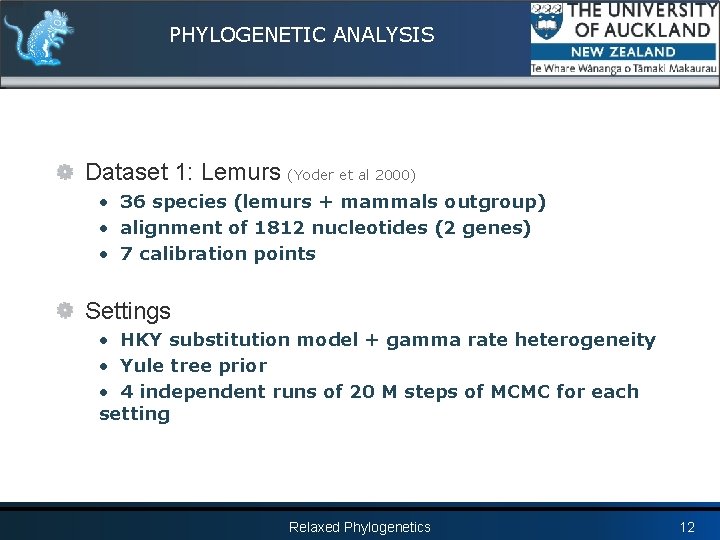
PHYLOGENETIC ANALYSIS Dataset 1: Lemurs (Yoder et al 2000) • 36 species (lemurs + mammals outgroup) • alignment of 1812 nucleotides (2 genes) • 7 calibration points Settings • HKY substitution model + gamma rate heterogeneity • Yule tree prior • 4 independent runs of 20 M steps of MCMC for each setting Relaxed Phylogenetics 12

PHYLOGENETIC ANALYSIS Dataset 2: Primates (Peter Waddell) • 7 species of primates: human, chimp, gorilla, orangutan, gibbon, macaque and marmoset • alignment of 1, 362, 261 nucleotides • Non coding regions • calibration : 16 MYA divergence time of human – orangutan Settings • GTR substitution model + gamma rate heterogeneity + Invariant • Coalescent or Yule tree prior • 4 independent runs of 50 M steps of MCMC for each setting Relaxed Phylogenetics 13

PHYLOGENETIC ANALYSIS Dataset 3: Yeast (Rokas et al 2003) • 8 species of yeast • alignment of 127, 026 nucleotides (106 genes) • calibration : Normal prior on the root height N (1, 0. 025) Settings • GTR substitution model + gamma rate heterogeneity + Invariant • Yule tree prior • 4 independent runs of 50 M steps of MCMC for each setting Relaxed Phylogenetics 14

PHYLOGENETIC ANALYSIS Dataset 4: Dengue (Rambaut 2000) • 17 serotype 4 sequences • alignment of 1, 485 nucleotides • serial sampling (1956 -1994) Settings • HKY substitution model • Coalescent tree prior • 4 independent runs of 10 M steps of MCMC for each setting Relaxed Phylogenetics 15

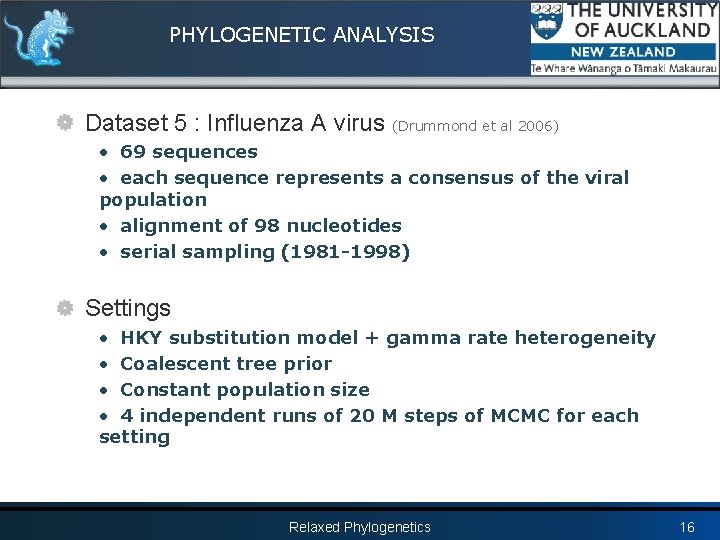
PHYLOGENETIC ANALYSIS Dataset 5 : Influenza A virus (Drummond et al 2006) • 69 sequences • each sequence represents a consensus of the viral population • alignment of 98 nucleotides • serial sampling (1981 -1998) Settings • HKY substitution model + gamma rate heterogeneity • Coalescent tree prior • Constant population size • 4 independent runs of 20 M steps of MCMC for each setting Relaxed Phylogenetics 16

MODEL COMPARISON Bayes Factor (Kass and Raftery 1995, Marc Suchard 2005) • Quantifies the real support of two competing hypothesis given the observed data → Ratio of the marginal likelihood of two models M 1 and M 2 → Bayesian analogue of the likelihood rate test (LRT) Relaxed Phylogenetics 17

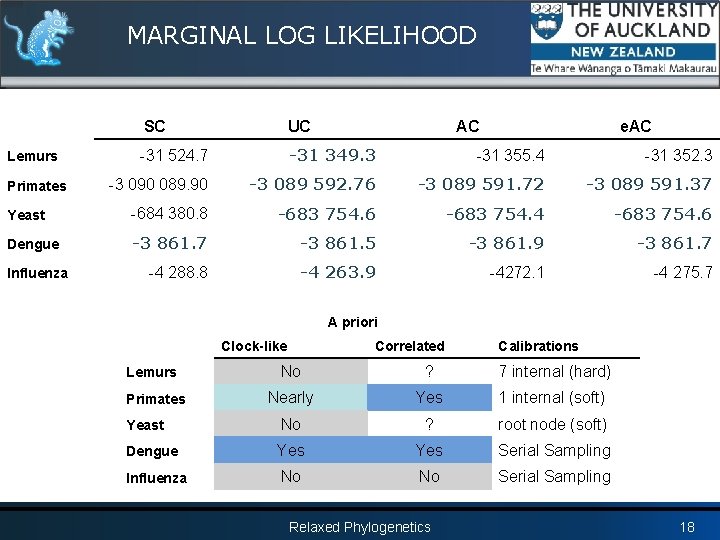
MARGINAL LOG LIKELIHOOD SC UC -31 524. 7 -31 349. 3 -31 355. 4 -31 352. 3 -3 090 089. 90 -3 089 592. 76 -3 089 591. 72 -3 089 591. 37 Yeast -684 380. 8 -683 754. 6 -683 754. 4 -683 754. 6 Dengue -3 861. 7 -3 861. 5 -3 861. 9 -3 861. 7 -4 288. 8 -4 263. 9 -4272. 1 -4 275. 7 Lemurs Primates Influenza AC e. AC A priori Clock-like Correlated Calibrations No ? 7 internal (hard) Nearly Yes 1 internal (soft) Yeast No ? root node (soft) Dengue Yes Serial Sampling Influenza No No Serial Sampling Lemurs Primates Relaxed Phylogenetics 18

Influenza dataset Consensus trees Uncorrelated Auto. Correlated Relaxed Phylogenetics 19

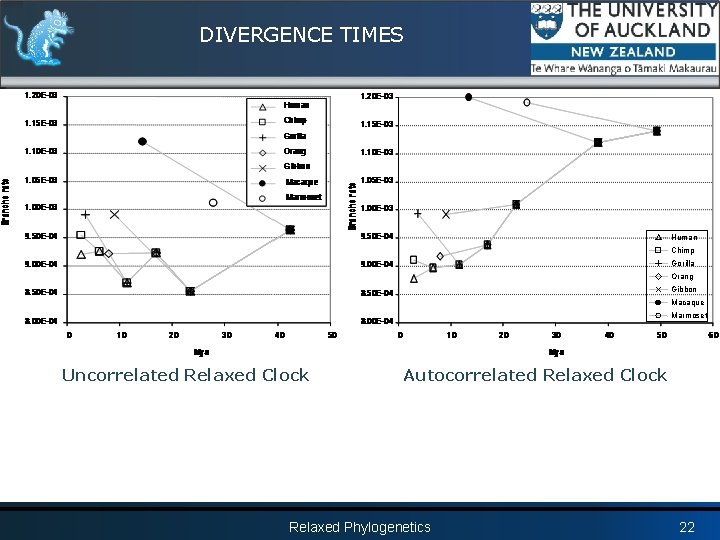
DIVERGENCE TIMES Relaxed Phylogenetics 20

DIVERGENCE TIMES Beast: mean of the posterior distributions, error bars are 95% lower and upper HPDs Glazko et al: error bars are +/- standard error Relaxed Phylogenetics 21

DIVERGENCE TIMES Human Chimp Gorilla Orang Gibbon Macaque Marmoset Uncorrelated Relaxed Clock Autocorrelated Relaxed Clock Relaxed Phylogenetics 22

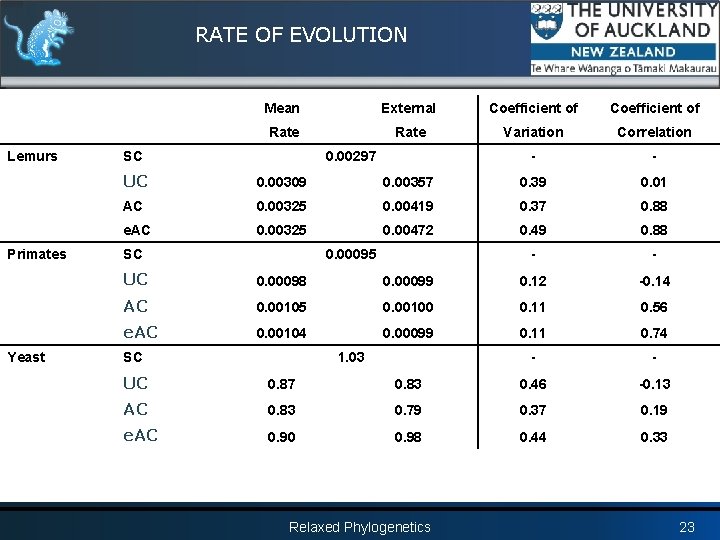
RATE OF EVOLUTION Lemurs Primates Yeast Mean External Coefficient of Rate Variation Correlation - - SC 0. 00297 UC 0. 00309 0. 00357 0. 39 0. 01 AC 0. 00325 0. 00419 0. 37 0. 88 e. AC 0. 00325 0. 00472 0. 49 0. 88 - - SC 0. 00095 UC 0. 00098 0. 00099 0. 12 -0. 14 AC 0. 00105 0. 00100 0. 11 0. 56 e. AC 0. 00104 0. 00099 0. 11 0. 74 - - SC 1. 03 UC 0. 87 0. 83 0. 46 -0. 13 AC 0. 83 0. 79 0. 37 0. 19 e. AC 0. 90 0. 98 0. 44 0. 33 Relaxed Phylogenetics 23

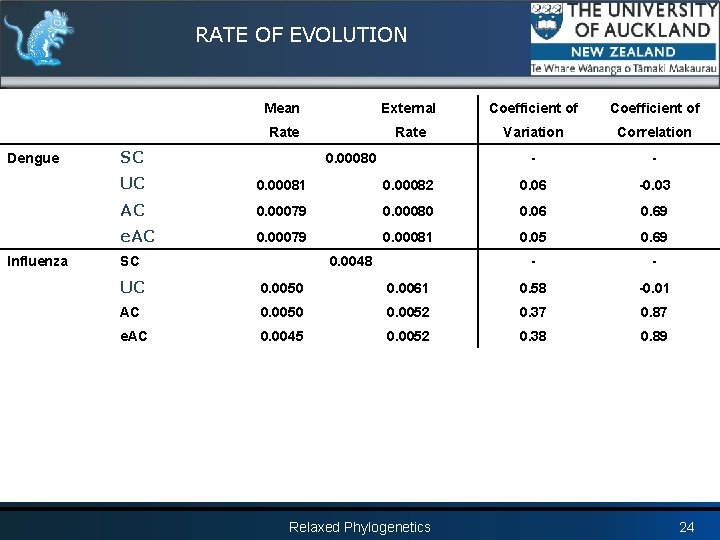
RATE OF EVOLUTION Dengue Influenza Mean External Coefficient of Rate Variation Correlation - - SC 0. 00080 UC 0. 00081 0. 00082 0. 06 -0. 03 AC 0. 00079 0. 00080 0. 06 0. 69 e. AC 0. 00079 0. 00081 0. 05 0. 69 - - SC 0. 0048 UC 0. 0050 0. 0061 0. 58 -0. 01 AC 0. 0050 0. 0052 0. 37 0. 87 e. AC 0. 0045 0. 0052 0. 38 0. 89 Relaxed Phylogenetics 24

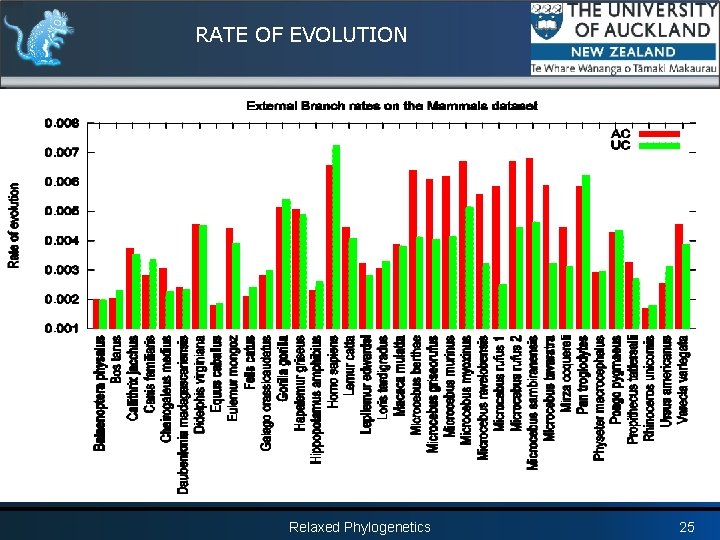
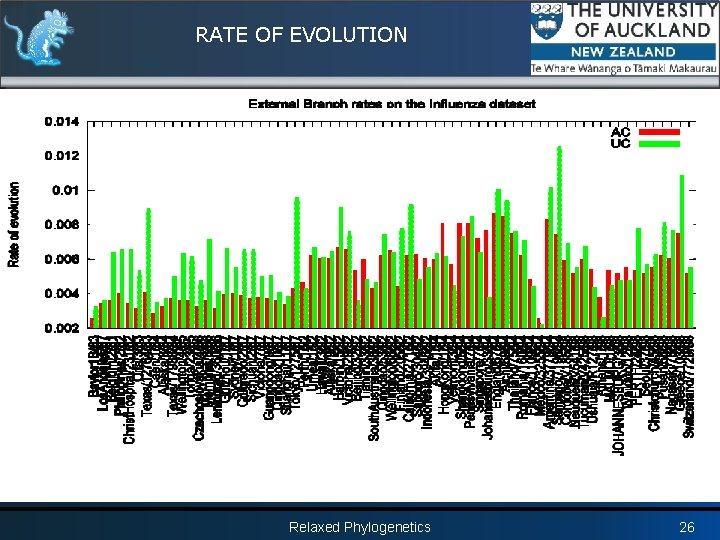
RATE OF EVOLUTION Relaxed Phylogenetics 25

RATE OF EVOLUTION Relaxed Phylogenetics 26

GENES RATE VS SPECIES RATE Mean rate per “locus” Primates Yeast Relaxed Phylogenetics 27

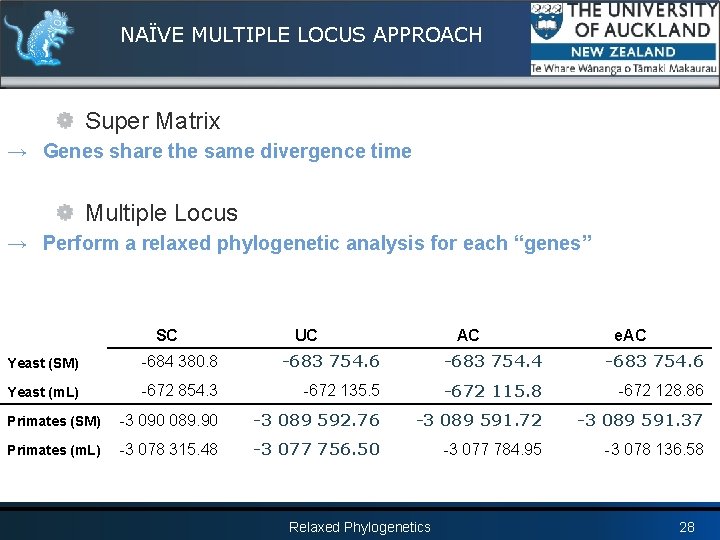
NAÏVE MULTIPLE LOCUS APPROACH Super Matrix → Genes share the same divergence time Multiple Locus → Perform a relaxed phylogenetic analysis for each “genes” SC UC AC e. AC Yeast (SM) -684 380. 8 -683 754. 6 -683 754. 4 -683 754. 6 Yeast (m. L) -672 854. 3 -672 135. 5 -672 115. 8 -672 128. 86 Primates (SM) -3 090 089. 90 -3 089 592. 76 -3 089 591. 72 -3 089 591. 37 Primates (m. L) -3 078 315. 48 -3 077 756. 50 -3 077 784. 95 -3 078 136. 58 Relaxed Phylogenetics 28

GENES DIVERGENCE TIMES VS SPECIES DIVERGENCE TIMES Relaxed Phylogenetics 29

GENES DIVERGENCE TIMES VS SPECIES DIVERGENCE TIMES Root Height in the primates dataset Relaxed Phylogenetics 30

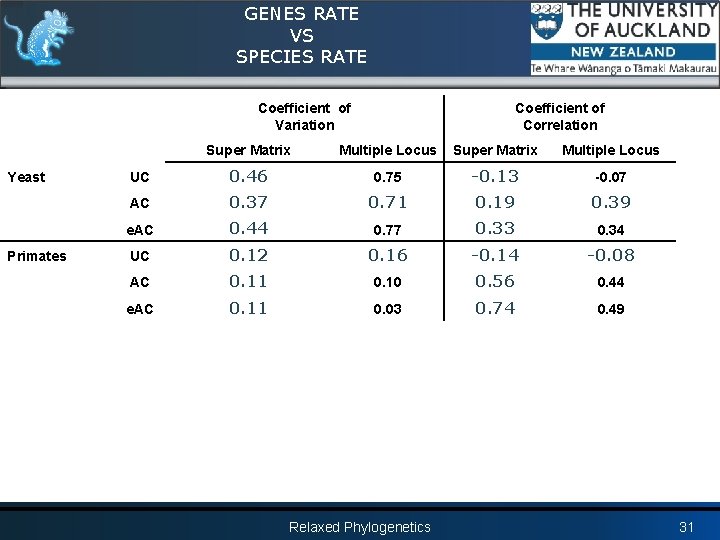
GENES RATE VS SPECIES RATE Coefficient of Variation Yeast Primates Coefficient of Correlation Super Matrix Multiple Locus UC 0. 46 0. 75 -0. 13 -0. 07 AC 0. 37 0. 71 0. 19 0. 39 e. AC 0. 44 0. 77 0. 33 0. 34 UC 0. 12 0. 16 -0. 14 -0. 08 AC 0. 11 0. 10 0. 56 0. 44 e. AC 0. 11 0. 03 0. 74 0. 49 Relaxed Phylogenetics 31

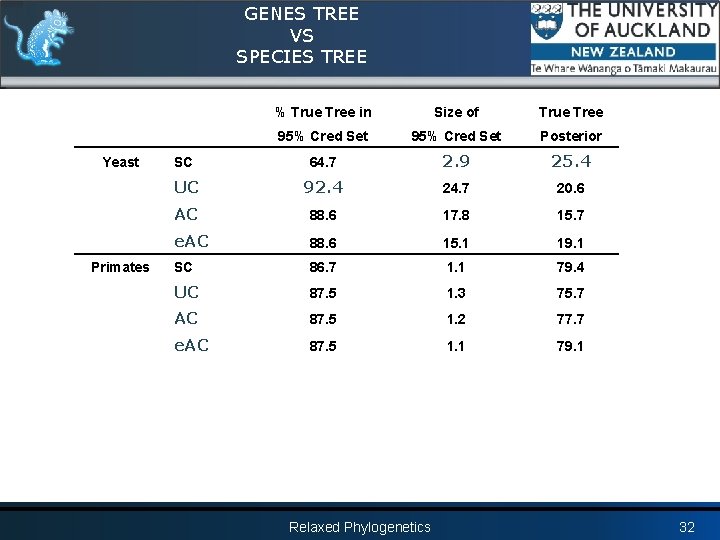
GENES TREE VS SPECIES TREE Yeast Primates % True Tree in Size of True Tree 95% Cred Set Posterior SC 64. 7 2. 9 25. 4 UC 92. 4 24. 7 20. 6 AC 88. 6 17. 8 15. 7 e. AC 88. 6 15. 1 19. 1 SC 86. 7 1. 1 79. 4 UC 87. 5 1. 3 75. 7 AC 87. 5 1. 2 77. 7 e. AC 87. 5 1. 1 79. 1 Relaxed Phylogenetics 32

GENES TREE VS SPECIES TREE Relaxed Phylogenetics 33

Conclusions Validation of the implementation in Beast Model comparison • • • Fit the data Uncorrelated vs Autocorrelated : prior knowledge Calibrations Estimate of rates Disagree in the multiple locus approach Reconstruct the tree topology Relaxed Phylogenetics 34

THANKS Relaxed Phylogenetics 35
 Taxonomic key for plants
Taxonomic key for plants Unscaled phylogenetic tree
Unscaled phylogenetic tree Mega phylogenetics
Mega phylogenetics Relaxed and stringent plasmid
Relaxed and stringent plasmid Principles of relaxed passive movements
Principles of relaxed passive movements Superlative of noisy
Superlative of noisy Relaxed and contracted sarcomere
Relaxed and contracted sarcomere 1/2 waist relaxed measurement
1/2 waist relaxed measurement Relaxed consistency
Relaxed consistency Relaxed work environment
Relaxed work environment A guest with relaxed inhibitions may
A guest with relaxed inhibitions may Kneel sitting muscle work
Kneel sitting muscle work Brutal denotation and connotation
Brutal denotation and connotation Slender connotation
Slender connotation Relaxed cone step mapping
Relaxed cone step mapping School nowadays
School nowadays Scary antonym
Scary antonym Michal cyprian
Michal cyprian Michal hammer
Michal hammer Michal lehnert
Michal lehnert Michal tkaczyk
Michal tkaczyk Michal simon
Michal simon Michal grešlik
Michal grešlik Michal kalman vek
Michal kalman vek Michal turošík
Michal turošík Michal drobot
Michal drobot Meniny michal
Meniny michal Michal bick
Michal bick Michal inbar-feigenberg
Michal inbar-feigenberg Michal kolcun
Michal kolcun Michal shinnar
Michal shinnar Inotaj v hadanke
Inotaj v hadanke Ttpet
Ttpet Hrábě skloňování vzor
Hrábě skloňování vzor Myš rod vzor
Myš rod vzor Michal tulek
Michal tulek