UltraHigh Resolution Xray Diffraction 12 Xray diffraction data







- Slides: 15

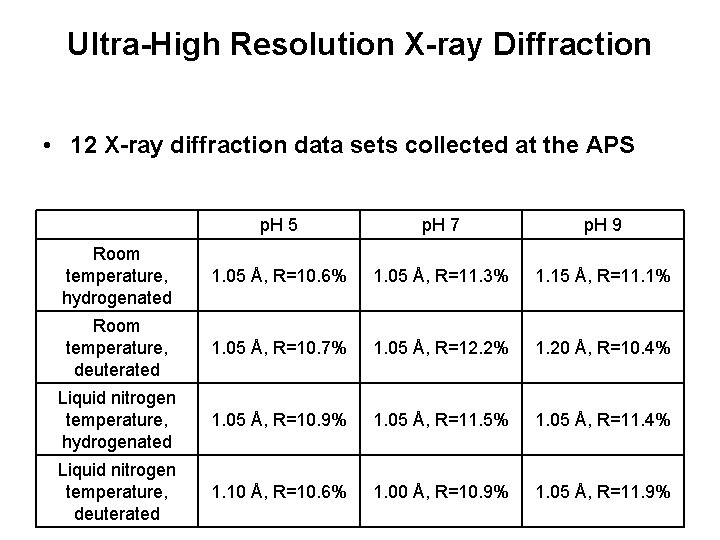
Ultra-High Resolution X-ray Diffraction • 12 X-ray diffraction data sets collected at the APS p. H 5 p. H 7 p. H 9 Room temperature, hydrogenated 1. 05 Å, R=10. 6% 1. 05 Å, R=11. 3% 1. 15 Å, R=11. 1% Room temperature, deuterated 1. 05 Å, R=10. 7% 1. 05 Å, R=12. 2% 1. 20 Å, R=10. 4% Liquid nitrogen temperature, hydrogenated 1. 05 Å, R=10. 9% 1. 05 Å, R=11. 5% 1. 05 Å, R=11. 4% Liquid nitrogen temperature, deuterated 1. 10 Å, R=10. 6% 1. 00 Å, R=10. 9% 1. 05 Å, R=11. 9%

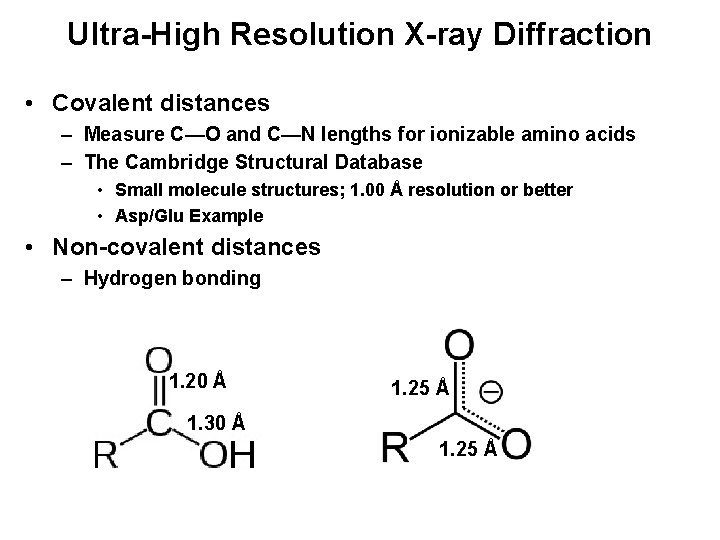
Ultra-High Resolution X-ray Diffraction • Covalent distances – Measure C—O and C—N lengths for ionizable amino acids – The Cambridge Structural Database • Small molecule structures; 1. 00 Å resolution or better • Asp/Glu Example • Non-covalent distances – Hydrogen bonding 1. 20 Å 1. 25 Å 1. 30 Å 1. 25 Å

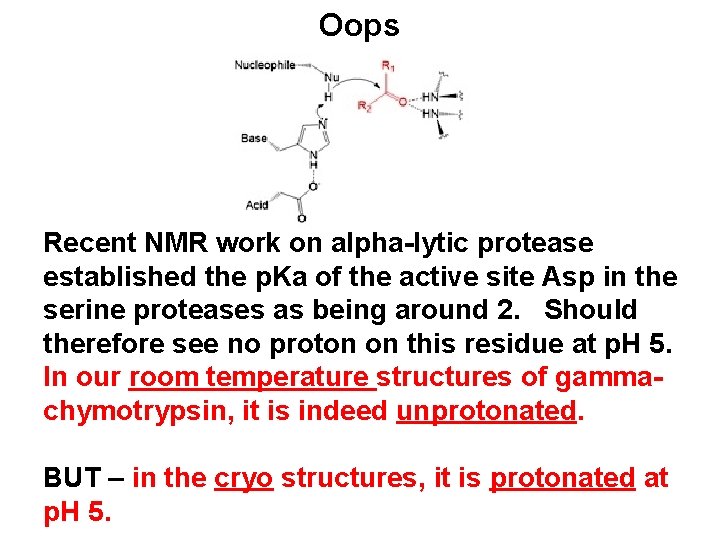
Oops Recent NMR work on alpha-lytic protease established the p. Ka of the active site Asp in the serine proteases as being around 2. Should therefore see no proton on this residue at p. H 5. In our room temperature structures of gammachymotrypsin, it is indeed unprotonated. BUT – in the cryo structures, it is protonated at p. H 5.

Precision and Accuracy of the Distance Data • Calculate RMSD of all bonds for individual residues • At highest resolution (1. 00 Å), differences greater than 0. 02 Å are significant • BUT – need “gold standard” where we are CERTAIN of the protonation state of all ionizable residues • SOLUTION – NEUTRON DIFFRACTION to locate deuterons directly from their nuclear density, at room temperature, at p. H 5, 7 and 9

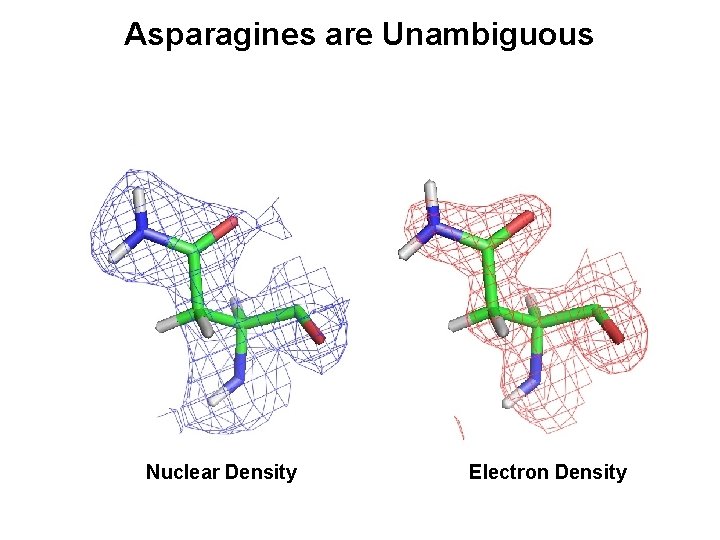
Asparagines are Unambiguous Nuclear Density Electron Density

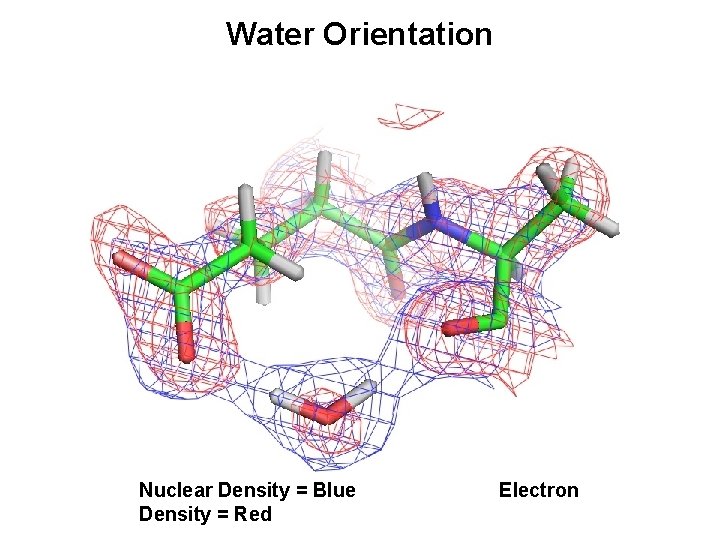
Water Orientation Nuclear Density = Blue Density = Red Electron

Structure 22, 899– 910, June 10, 2014


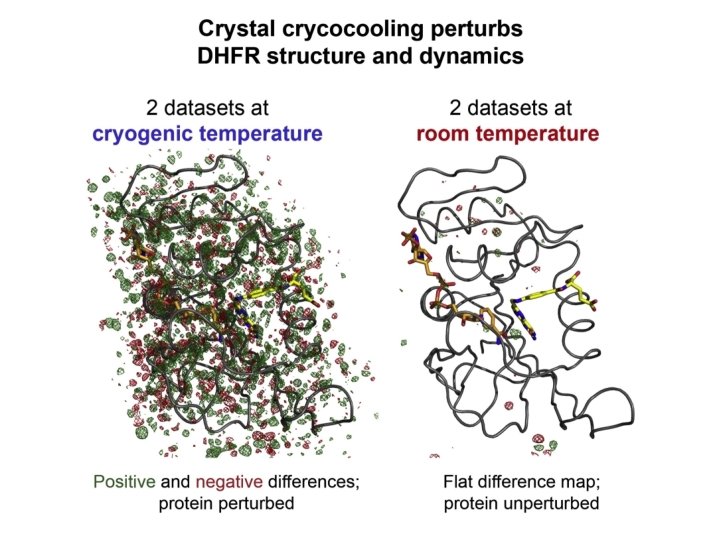
To Summarize • Nearly all the structures in the Protein Data Bank in the last 25 -30 years have been determined at -200 o. C, more than 100 degrees below the glass transition in protein dynamics • Such low temperatures have clear-cut effects on protein dynamics and protein volume and internal packing, but indeterminate effects on loop and side-chain conformations, interdomain positions, residue protonation states, and metal ion spin states, among other things

A Proposal The protein crystallographic community should make a concerted effort to determine at least one room temperature crystal structure for every unique protein in the Protein Data Bank, at the highest possible resolution

A Proposal The protein crystallographic community should make a concerted effort to determine at least one room temperature crystal structure for every unique protein in the Protein Data Bank, at the highest possible resolution. This might be a very good use for Free Electron Laser crystallography




Tom Alber