Typical Aligned Biological Sequence Data MAXmouse DMAXMOUSE MAX
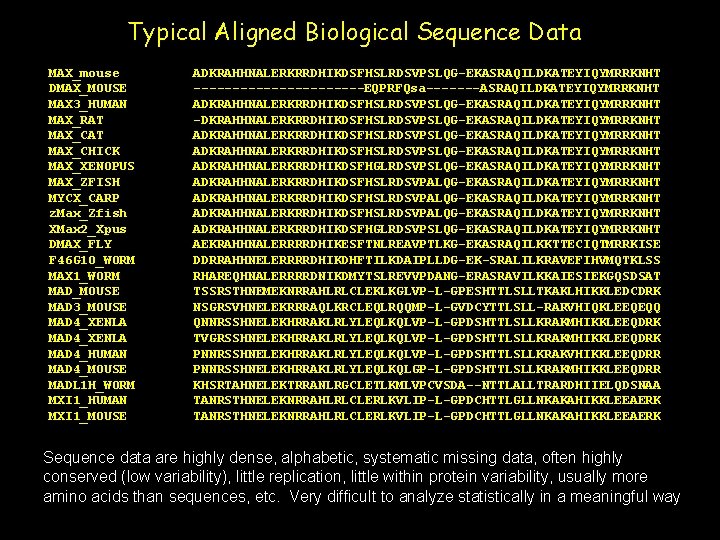
Typical Aligned Biological Sequence Data MAX_mouse DMAX_MOUSE MAX 3_HUMAN MAX_RAT MAX_CHICK MAX_XENOPUS MAX_ZFISH MYCX_CARP z. Max_Zfish XMax 2_Xpus DMAX_FLY F 46 G 10_WORM MAX 1_WORM MAD_MOUSE MAD 3_MOUSE MAD 4_XENLA MAD 4_HUMAN MAD 4_MOUSE MADL 1 H_WORM MXI 1_HUMAN MXI 1_MOUSE ADKRAHHNALERKRRDHIKDSFHSLRDSVPSLQG-EKASRAQILDKATEYIQYMRRKNHT -----------EQPRFQsa-------ASRAQILDKATEYIQYMRRKNHT ADKRAHHNALERKRRDHIKDSFHSLRDSVPSLQG-EKASRAQILDKATEYIQYMRRKNHT -DKRAHHNALERKRRDHIKDSFHSLRDSVPSLQG-EKASRAQILDKATEYIQYMRRKNHT ADKRAHHNALERKRRDHIKDSFHSLRDSVPSLQG-EKASRAQILDKATEYIQYMRRKNHT ADKRAHHNALERKRRDHIKDSFHGLRDSVPSLQG-EKASRAQILDKATEYIQYMRRKNHT ADKRAHHNALERKRRDHIKDSFHSLRDSVPALQG-EKASRAQILDKATEYIQYMRRKNHT ADKRAHHNALERKRRDHIKDSFHGLRDSVPSLQG-EKASRAQILDKATEYIQYMRRKNHT AEKRAHHNALERRRRDHIKESFTNLREAVPTLKG-EKASRAQILKKTTECIQTMRRKISE DDRRAHHNELERRRRDHIKDHFTILKDAIPLLDG-EK-SRALILKRAVEFIHVMQTKLSS RHAREQHNALERRRRDNIKDMYTSLREVVPDANG-ERASRAVILKKAIESIEKGQSDSAT TSSRSTHNEMEKNRRAHLRLCLEKLKGLVP-L-GPESHTTLSLLTKAKLHIKKLEDCDRK NSGRSVHNELEKRRRAQLKRCLEQLRQQMP-L-GVDCYTTLSLL-RARVHIQKLEEQEQQ QNNRSSHNELEKHRRAKLRLYLEQLKQLVP-L-GPDSHTTLSLLKRAKMHIKKLEEQDRK TVGRSSHNELEKHRRAKLRLYLEQLKQLVP-L-GPDSHTTLSLLKRAKMHIKKLEEQDRK PNNRSSHNELEKHRRAKLRLYLEQLKQLVP-L-GPDSHTTLSLLKRAKVHIKKLEEQDRR PNNRSSHNELEKHRRAKLRLYLEQLKQLGP-L-GPDSHTTLSLLKRAKMHIKKLEEQDRR KHSRTAHNELEKTRRANLRGCLETLKMLVPCVSDA--NTTLALLTRARDHIIELQDSNAA TANRSTHNELEKNRRAHLRLCLERLKVLIP-L-GPDCHTTLGLLNKAKAHIKKLEEAERK Sequence data are highly dense, alphabetic, systematic missing data, often highly conserved (low variability), little replication, little within protein variability, usually more amino acids than sequences, etc. Very difficult to analyze statistically in a meaningful way
A major goal of biological research is to provide general models or systematic principals to explain complex phenomena. In proteins, this involves the molecular architecture of their component parts their origins, regulation, interrelationships and evolution.
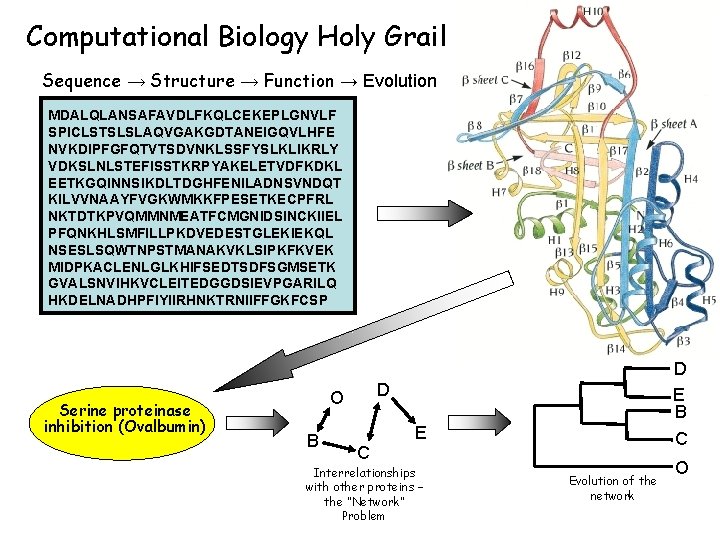
Computational Biology Holy Grail Sequence → Structure → Function → Evolution MDALQLANSAFAVDLFKQLCEKEPLGNVLF SPICLSTSLSLAQVGAKGDTANEIGQVLHFE NVKDIPFGFQTVTSDVNKLSSFYSLKLIKRLY VDKSLNLSTEFISSTKRPYAKELETVDFKDKL EETKGQINNSIKDLTDGHFENILADNSVNDQT KILVVNAAYFVGKWMKKFPESETKECPFRL NKTDTKPVQMMNMEATFCMGNIDSINCKIIEL PFQNKHLSMFILLPKDVEDESTGLEKIEKQL NSESLSQWTNPSTMANAKVKLSIPKFKVEK MIDPKACLENLGLKHIFSEDTSDFSGMSETK GVALSNVIHKVCLEITEDGGDSIEVPGARILQ HKDELNADHPFIYIIRHNKTRNIIFFGKFCSP Serine proteinase inhibition (Ovalbumin) D O B D E B C E Interrelationships with other proteins – the “Network” Problem C Evolution of the network O

Are phylogenetic trees the best way to describe protein sequence variability and evolution?

Phylogenetic Trees: Pros Trees are good for • describing hierarchical patterns • clustering taxa • describing extent of lineage divergence • estimating ancestral relationships • summarizing overall changes in data.

Phylogenetic Trees: cons Trees are NOT very good for: • understanding dimensionality • analyzing covariation • describing the biological basis of evolutionary divergence • elucidating the components upon which selection might operate
Significance of Covariation • Understanding covariation is fundamental to modeling protein structure and evolution • Evolutionary and structural change constrained by covariation among amino acids • Accurate prediction of protein structure requires knowledge of covariance structure • Covariation reduces the dimensionality of phylogenetic information • Analytical procedures (like ML) make strong assumptions about covariances
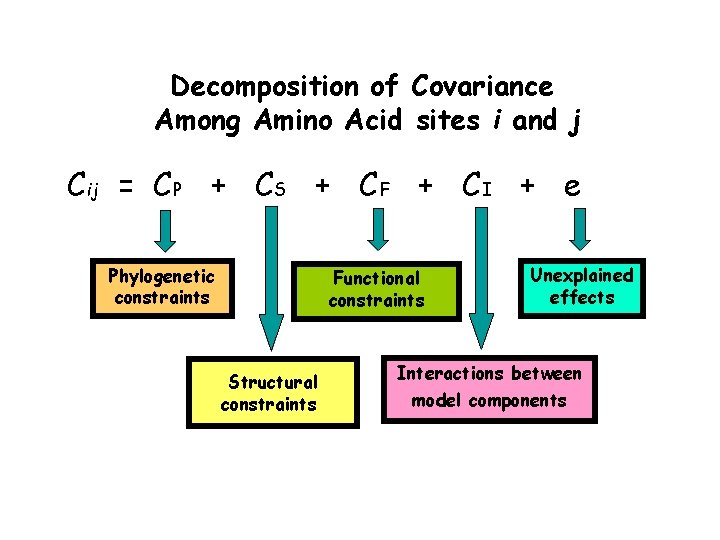
Decomposition of Covariance Among Amino Acid sites i and j Cij = CP + CS + CF + CI + e Phylogenetic constraints Functional constraints Structural constraints Unexplained effects Interactions between model components
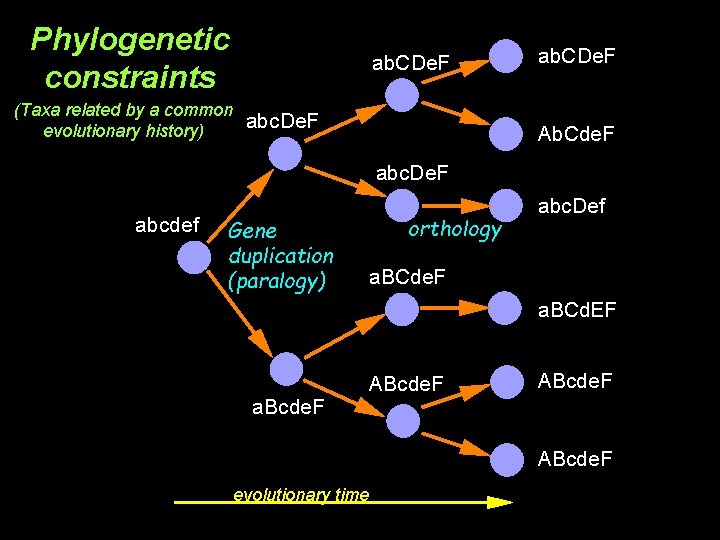
Phylogenetic constraints ab. CDe. F (Taxa related by a common abc. De. F evolutionary history) ab. CDe. F Ab. Cde. F abc. De. F abcdef Gene duplication (paralogy) orthology abc. Def a. BCde. F a. BCd. EF a. Bcde. F ABcde. F evolutionary time
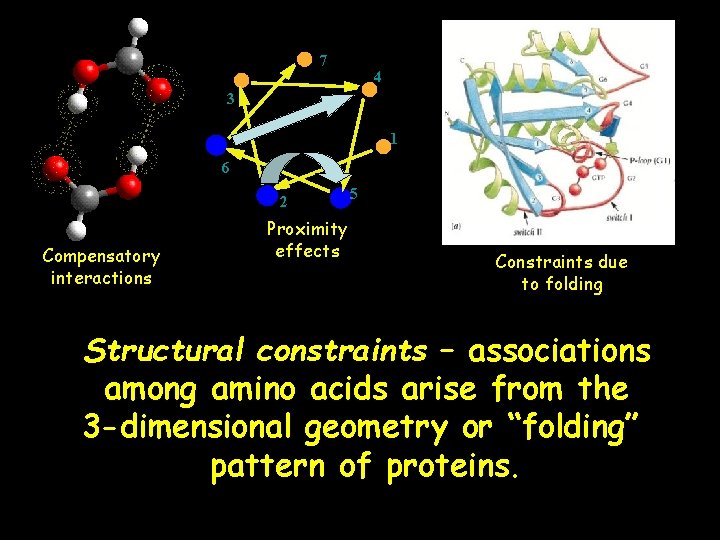
7 4 3 1 6 2 Compensatory interactions Proximity effects 5 Constraints due to folding Structural constraints – associations among amino acids arise from the 3 -dimensional geometry or “folding” pattern of proteins.

Some Structural Associations in Proteins Hydrophobic interactions: associations among amino acids with non-polar side chains Salt bridges: correlations between charged residues Hydrogen bonds: correlations between electron donors and receptors Size constraints: correlations between large and small side chains
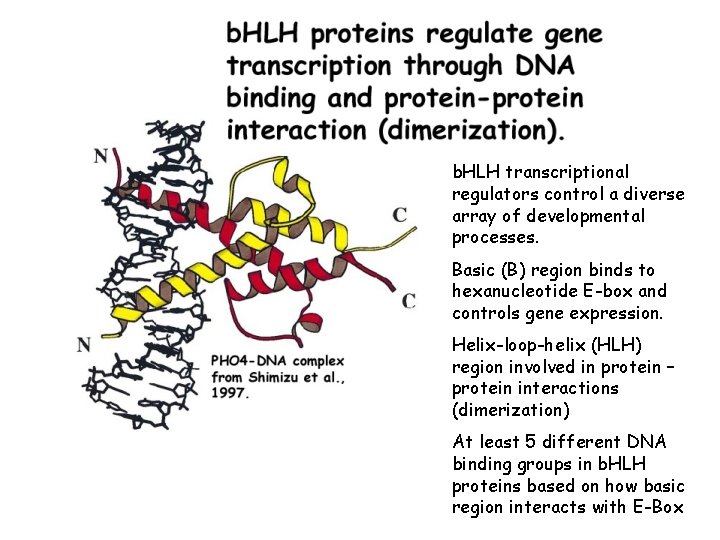
b. HLH transcriptional regulators control a diverse array of developmental processes. Basic (B) region binds to hexanucleotide E-box and controls gene expression. Helix-loop-helix (HLH) region involved in protein – protein interactions (dimerization) At least 5 different DNA binding groups in b. HLH proteins based on how basic region interacts with E-Box
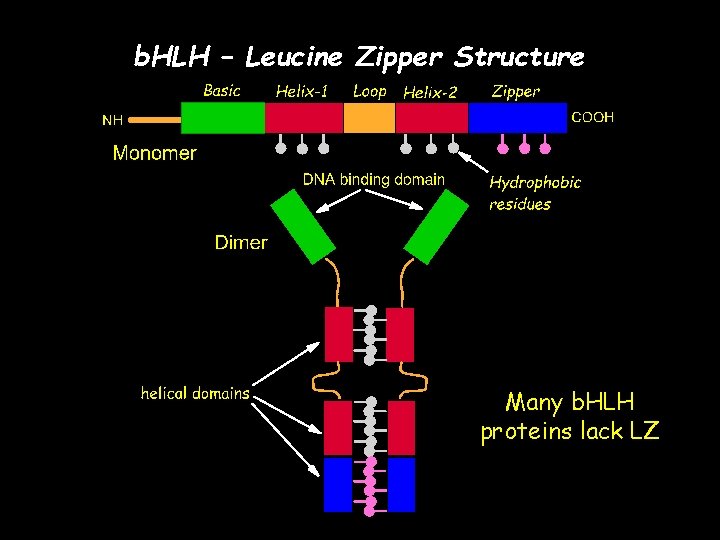
b. HLH – Leucine Zipper Structure Many b. HLH proteins lack LZ
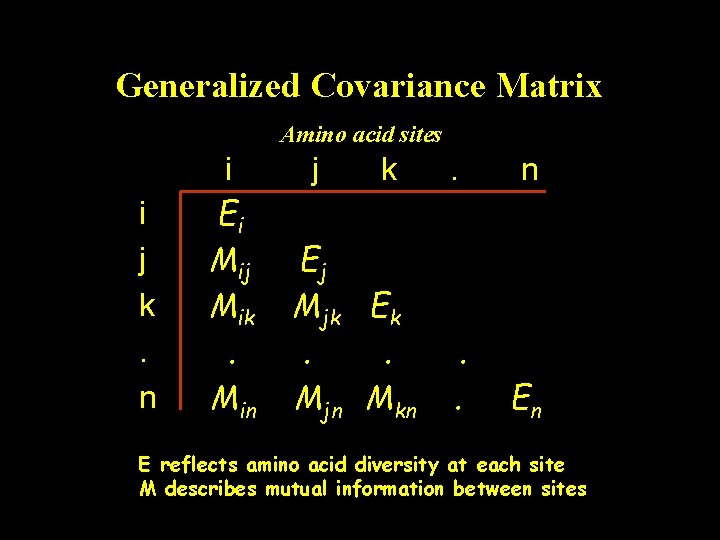
Generalized Covariance Matrix Amino acid sites i j k. n i Ei Mij Mik. Min j k Ej Mjk Ek. . Mjn Mkn . . En E reflects amino acid diversity at each site M describes mutual information between sites
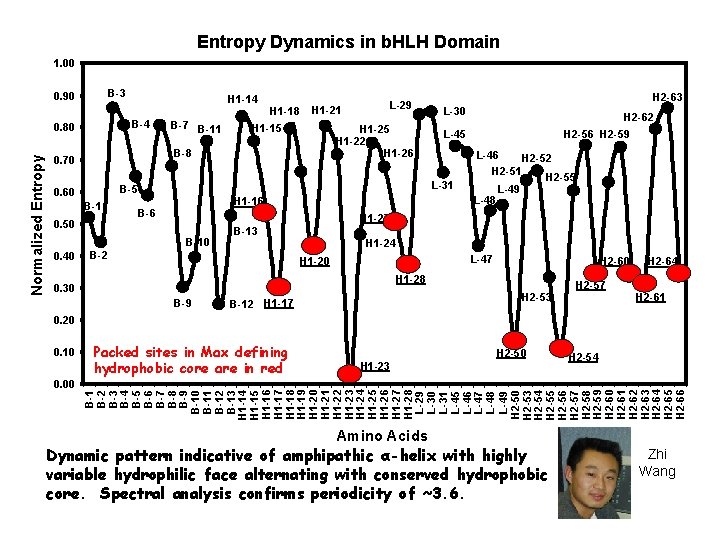
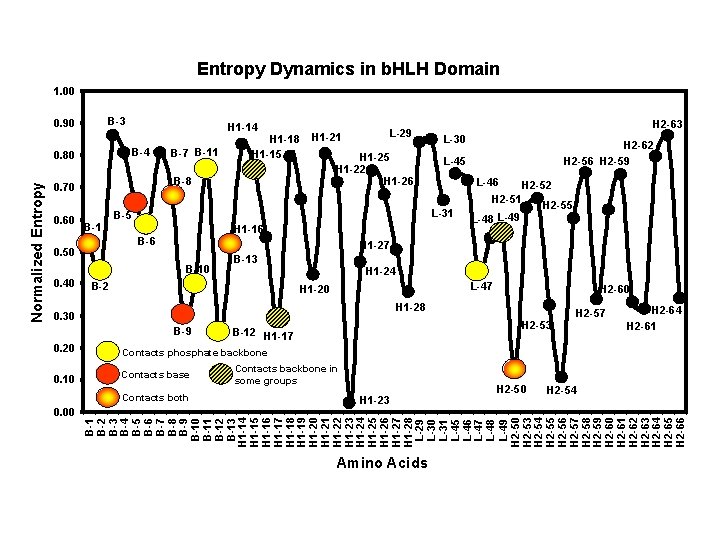
Entropy Dynamics in b. HLH Domain 1. 00 B-3 0. 90 B-4 0. 80 B-7 B-11 H 1 -15 H 1 -22 H 1 -26 B-1 0. 50 H 2 -63 L-30 H 1 -16 B-6 H 2 -62 L-45 L-31 B-5 0. 60 L-29 H 1 -21 B-8 0. 70 H 2 -56 H 2 -59 L-46 H 2 -52 H 2 -51 H 2 -55 L-49 L-48 H 1 -27 B-10 0. 40 H 1 -18 B-13 B-2 H 1 -24 L-47 H 1 -20 H 2 -60 H 1 -28 0. 30 B-9 H 2 -53 B-12 H 1 -17 H 2 -57 H 2 -64 H 2 -61 0. 20 0. 10 0. 00 Packed sites in Max defining hydrophobic core are in red H 2 -50 H 1 -23 H 2 -54 B-1 B-2 B-3 B-4 B-5 B-6 B-7 B-8 B-9 B-10 B-11 B-12 B-13 H 1 -14 H 1 -15 H 1 -16 H 1 -17 H 1 -18 H 1 -19 H 1 -20 H 1 -21 H 1 -22 H 1 -23 H 1 -24 H 1 -25 H 1 -26 H 1 -27 H 1 -28 L-29 L-30 L-31 L-45 L-46 L-47 L-48 L-49 H 2 -50 H 2 -53 H 2 -54 H 2 -55 H 2 -56 H 2 -57 H 2 -58 H 2 -59 H 2 -60 H 2 -61 H 2 -62 H 2 -63 H 2 -64 H 2 -65 H 2 -66 Normalized Entropy H 1 -14 Amino Acids Dynamic pattern indicative of amphipathic α-helix with highly variable hydrophilic face alternating with conserved hydrophobic core. Spectral analysis confirms periodicity of ~3. 6. Zhi Wang
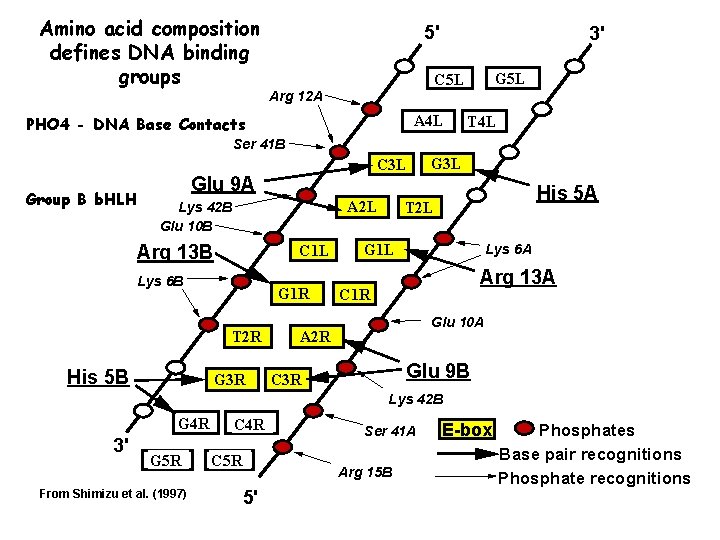
Amino acid composition defines DNA binding groups 5' 3' G 5 L C 5 L Arg 12 A A 4 L PHO 4 - DNA Base Contacts T 4 L Ser 41 B Group B b. HLH C 3 L Glu 9 A A 2 L Lys 42 B Glu 10 B Arg 13 B C 1 L Lys 6 B G 1 R T 2 R His 5 B G 3 R G 3 L His 5 A T 2 L Lys 6 A G 1 L Arg 13 A C 1 R Glu 10 A A 2 R Glu 9 B C 3 R Lys 42 B G 4 R 3' G 5 R From Shimizu et al. (1997) C 4 R C 5 R Ser 41 A Arg 15 B 5' E-box Phosphates Base pair recognitions Phosphate recognitions
Entropy Dynamics in b. HLH Domain 1. 00 B-3 0. 90 B-4 0. 80 B-1 H 1 -16 B-2 H 2 -56 H 2 -59 L-46 H 2 -52 H 2 -51 H 2 -55 L-48 L-49 L-47 H 2 -60 H 1 -28 H 2 -53 B-12 H 1 -17 H 2 -57 H 2 -64 H 2 -61 Contacts phosphate backbone Contacts base Contacts backbone in some groups both Packed Contacts sites in Max are underlined 0. 00 L-45 H 1 -24 H 1 -20 B-9 0. 10 H 2 -62 H 1 -27 B-13 0. 30 0. 20 H 2 -63 L-30 L-31 B-10 0. 40 H 1 -25 H 1 -22 H 1 -26 B-5 B-6 0. 50 L-29 H 1 -21 B-8 0. 70 0. 60 B-7 B-11 H 1 -18 H 1 -15 H 1 -23 H 2 -50 H 2 -54 B-1 B-2 B-3 B-4 B-5 B-6 B-7 B-8 B-9 B-10 B-11 B-12 B-13 H 1 -14 H 1 -15 H 1 -16 H 1 -17 H 1 -18 H 1 -19 H 1 -20 H 1 -21 H 1 -22 H 1 -23 H 1 -24 H 1 -25 H 1 -26 H 1 -27 H 1 -28 L-29 L-30 L-31 L-45 L-46 L-47 L-48 L-49 H 2 -50 H 2 -53 H 2 -54 H 2 -55 H 2 -56 H 2 -57 H 2 -58 H 2 -59 H 2 -60 H 2 -61 H 2 -62 H 2 -63 H 2 -64 H 2 -65 H 2 -66 Normalized Entropy H 1 -14 Amino Acids
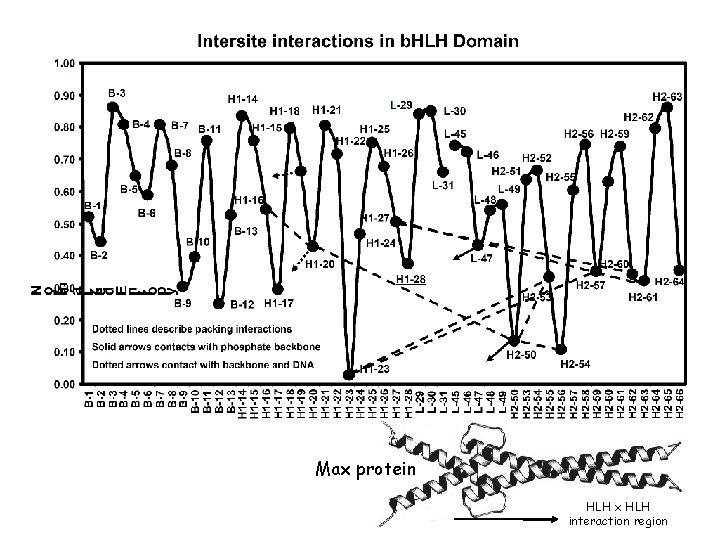
Max protein HLH x HLH interaction region
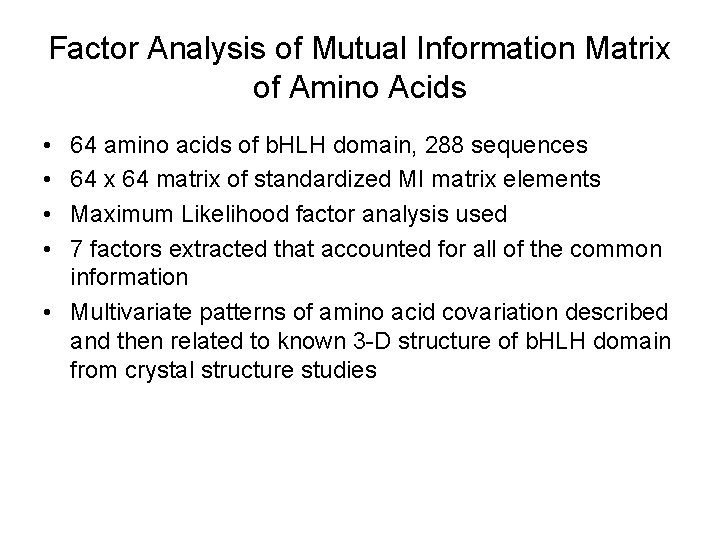
Factor Analysis of Mutual Information Matrix of Amino Acids • • 64 amino acids of b. HLH domain, 288 sequences 64 x 64 matrix of standardized MI matrix elements Maximum Likelihood factor analysis used 7 factors extracted that accounted for all of the common information • Multivariate patterns of amino acid covariation described and then related to known 3 -D structure of b. HLH domain from crystal structure studies
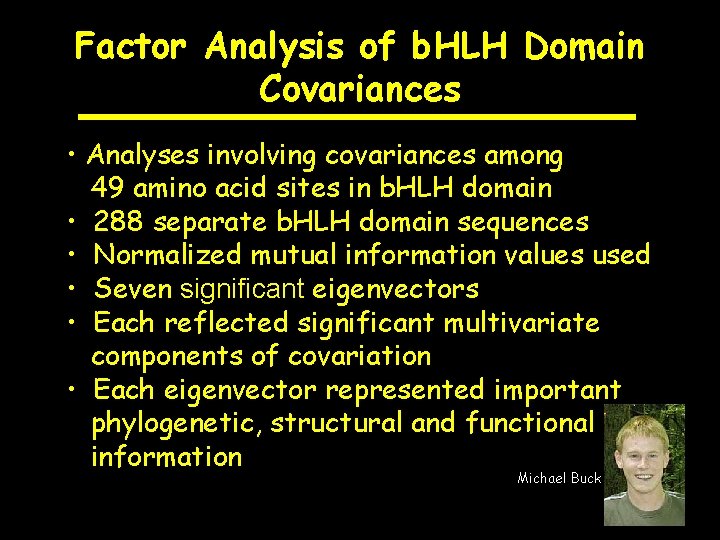
Factor Analysis of b. HLH Domain Covariances • Analyses involving covariances among 49 amino acid sites in b. HLH domain • 288 separate b. HLH domain sequences • Normalized mutual information values used • Seven significant eigenvectors • Each reflected significant multivariate components of covariation • Each eigenvector represented important phylogenetic, structural and functional information Michael Buck
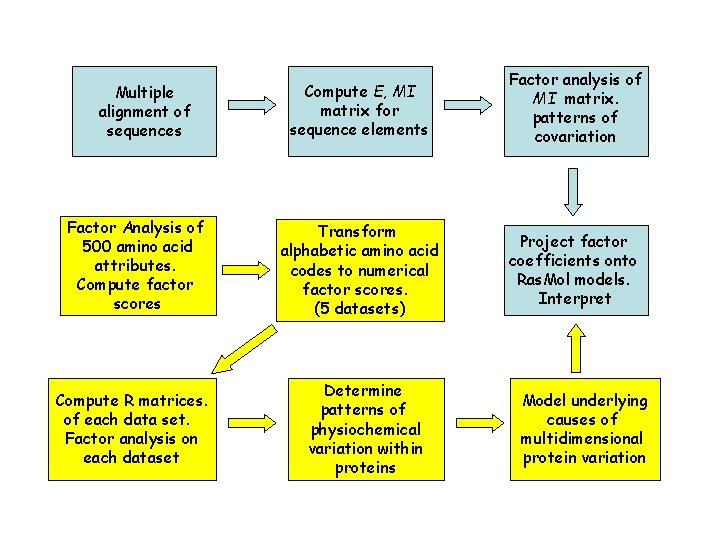
Flow of Statistical Analyses Multiple alignment of sequences Factor Analysis of 500 amino acid attributes. Compute factor scores Compute R matrices. of each data set. Factor analysis on each dataset Compute E, MI matrix for sequence elements Factor analysis of MI matrix. patterns of covariation Transform alphabetic amino acid codes to numerical factor scores. (5 datasets) Project factor coefficients onto Ras. Mol models. Interpret Determine patterns of physiochemical variation within proteins Model underlying causes of multidimensional protein variation
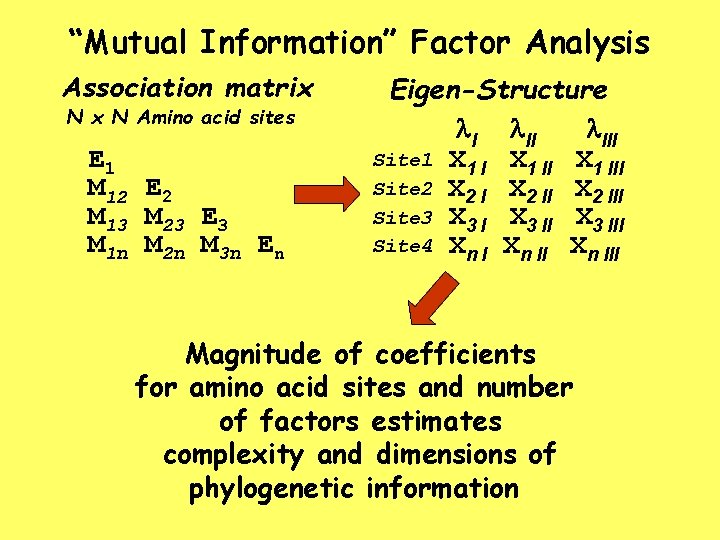
“Mutual Information” Factor Analysis Association matrix N Amino acid sites E 1 M 12 E 2 M 13 M 23 E 3 M 1 n M 2 n M 3 n En Eigen-Structure l. III Site 1 X 1 III Site 2 X 2 III Site 3 X 3 III Site 4 Xn III Magnitude of coefficients for amino acid sites and number of factors estimates complexity and dimensions of phylogenetic information
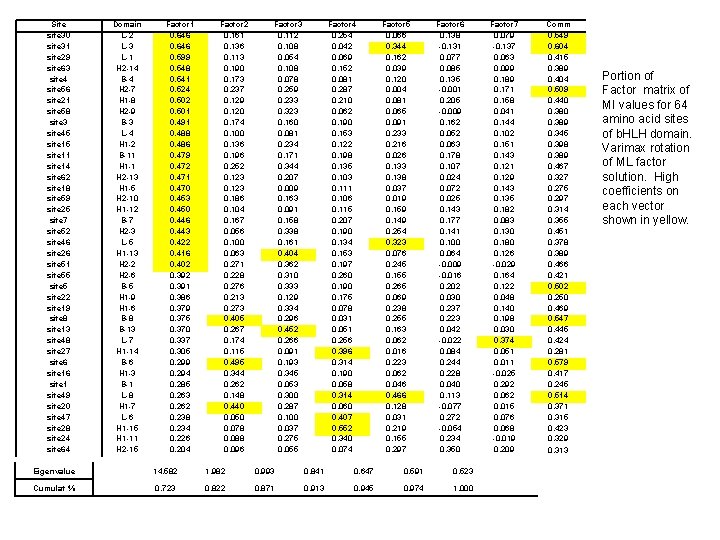
Site site 30 site 31 site 29 site 63 site 4 site 56 site 21 site 58 site 3 site 45 site 11 site 14 site 62 site 18 site 59 site 25 site 7 site 52 site 46 site 26 site 51 site 55 site 22 site 19 site 8 site 13 site 48 site 27 site 6 site 1 site 49 site 20 site 47 site 28 site 24 site 64 Domain L-2 L-3 L-1 H 2 -14 B-4 H 2 -7 H 1 -8 H 2 -9 B-3 L-4 H 1 -2 B-11 H 1 -1 H 2 -13 H 1 -5 H 2 -10 H 1 -12 B-7 H 2 -3 L-5 H 1 -13 H 2 -2 H 2 -6 B-5 H 1 -9 H 1 -6 B-8 B-13 L-7 H 1 -14 B-6 H 1 -3 B-1 L-8 H 1 -7 L-6 H 1 -15 H 1 -11 H 2 -15 Factor 1 0. 646 0. 599 0. 548 0. 541 0. 524 0. 502 0. 501 0. 491 0. 488 0. 486 0. 479 0. 472 0. 471 0. 470 0. 453 0. 450 0. 446 0. 443 0. 422 0. 416 0. 402 0. 391 0. 386 0. 379 0. 375 0. 370 0. 337 0. 305 0. 299 0. 294 0. 285 0. 263 0. 262 0. 238 0. 234 0. 226 0. 204 Factor 2 0. 161 0. 136 0. 113 0. 190 0. 173 0. 237 0. 129 0. 120 0. 174 0. 100 0. 136 0. 196 0. 252 0. 123 0. 186 0. 104 0. 167 0. 056 0. 100 0. 063 0. 271 0. 228 0. 276 0. 213 0. 273 0. 405 0. 267 0. 174 0. 115 0. 495 0. 344 0. 262 0. 148 0. 440 0. 050 0. 078 0. 088 0. 096 Factor 3 0. 112 0. 108 0. 054 0. 108 0. 078 0. 259 0. 233 0. 323 0. 160 0. 081 0. 234 0. 171 0. 344 0. 207 0. 009 0. 163 0. 091 0. 158 0. 338 0. 161 0. 404 0. 362 0. 310 0. 333 0. 129 0. 334 0. 296 0. 452 0. 266 0. 091 0. 193 0. 345 0. 053 0. 300 0. 287 0. 100 0. 037 0. 275 0. 055 Factor 4 0. 254 0. 042 0. 069 0. 152 0. 081 0. 287 0. 210 0. 062 0. 190 0. 153 0. 122 0. 198 0. 135 0. 103 0. 111 0. 106 0. 115 0. 207 0. 190 0. 134 0. 153 0. 197 0. 260 0. 190 0. 175 0. 078 0. 031 0. 051 0. 256 0. 396 0. 314 0. 190 0. 058 0. 314 0. 060 0. 407 0. 552 0. 340 0. 074 Factor 5 0. 066 0. 344 0. 162 0. 039 0. 120 0. 004 0. 081 0. 065 0. 091 0. 233 0. 216 0. 026 0. 133 0. 138 0. 037 0. 019 0. 159 0. 149 0. 254 0. 323 0. 076 0. 245 0. 155 0. 265 0. 069 0. 238 0. 255 0. 163 0. 062 0. 016 0. 223 0. 062 0. 046 0. 466 0. 128 0. 031 0. 219 0. 155 0. 297 Factor 6 0. 138 -0. 131 0. 077 0. 085 0. 135 -0. 001 0. 205 -0. 009 0. 162 0. 052 0. 063 0. 178 0. 107 0. 024 0. 072 0. 025 0. 143 0. 177 0. 141 0. 100 0. 064 -0. 009 -0. 016 0. 202 0. 030 0. 237 0. 223 0. 042 -0. 022 0. 084 0. 244 0. 228 0. 040 0. 113 -0. 077 0. 272 -0. 054 0. 234 0. 350 Eigenvalue 14. 582 1. 982 0. 993 0. 841 0. 647 0. 591 0. 523 Cumulat % 0. 723 0. 822 0. 871 0. 913 0. 945 0. 974 1. 000 Factor 7 0. 079 -0. 137 0. 063 0. 099 0. 189 0. 171 0. 158 0. 041 0. 144 0. 102 0. 151 0. 143 0. 121 0. 129 0. 143 0. 135 0. 182 0. 083 0. 130 0. 180 0. 126 -0. 029 0. 164 0. 122 0. 048 0. 140 0. 198 0. 030 0. 374 0. 051 0. 011 -0. 025 0. 292 0. 062 0. 015 0. 076 0. 068 -0. 019 0. 209 Comm 0. 549 0. 604 0. 415 0. 389 0. 404 0. 509 0. 440 0. 389 0. 345 0. 398 0. 389 0. 467 0. 327 0. 275 0. 297 0. 314 0. 355 0. 451 0. 378 0. 389 0. 466 0. 421 0. 502 0. 250 0. 469 0. 547 0. 445 0. 424 0. 281 0. 579 0. 417 0. 245 0. 514 0. 371 0. 315 0. 423 0. 329 0. 313 Portion of Factor matrix of MI values for 64 amino acid sites of b. HLH domain. Varimax rotation of ML factor solution. High coefficients on each vector shown in yellow.

Factor analyses describe: • Dimensionality of shared and unique covariation • Major patterns of amino acid covariation among all major b. HLH lineages • A model for structural and sequence evolution in b. HLH • An understanding of the biological bases of simultaneous changes among amino acid sites
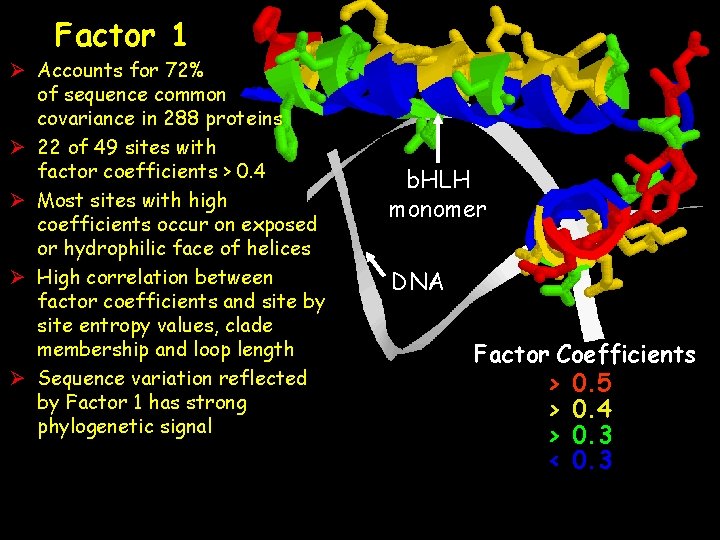
Factor 1 Ø Accounts for 72% of sequence common covariance in 288 proteins Ø 22 of 49 sites with factor coefficients > 0. 4 Ø Most sites with high coefficients occur on exposed or hydrophilic face of helices Ø High correlation between factor coefficients and site by site entropy values, clade membership and loop length Ø Sequence variation reflected by Factor 1 has strong phylogenetic signal b. HLH monomer DNA Factor Coefficients > 0. 5 > 0. 4 > 0. 3 < 0. 3
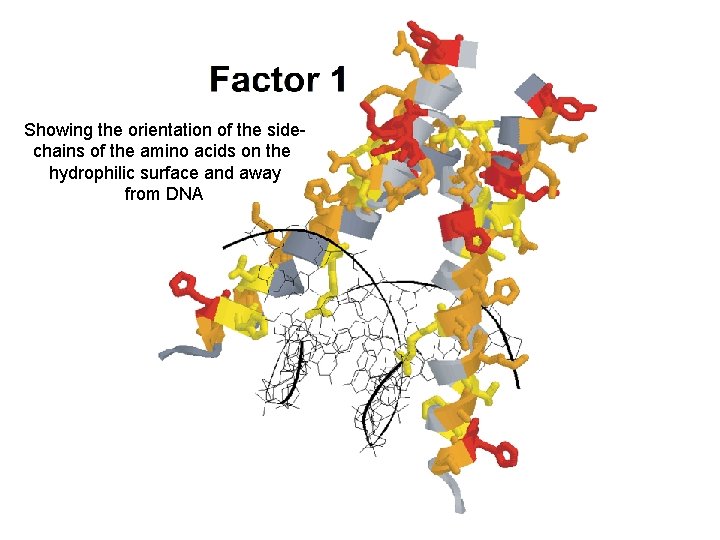
Showing the orientation of the sidechains of the amino acids on the hydrophilic surface and away from DNA
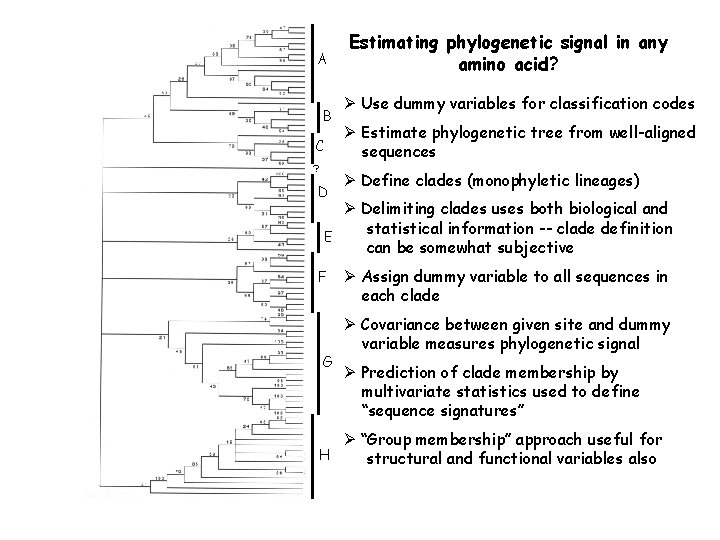
A B C ? D Estimating phylogenetic signal in any amino acid? Ø Use dummy variables for classification codes Ø Estimate phylogenetic tree from well-aligned sequences Ø Define clades (monophyletic lineages) Ø Delimiting clades uses both biological and statistical information -- clade definition E can be somewhat subjective F G H Ø Assign dummy variable to all sequences in each clade Ø Covariance between given site and dummy variable measures phylogenetic signal Ø Prediction of clade membership by multivariate statistics used to define “sequence signatures” Ø “Group membership” approach useful for structural and functional variables also
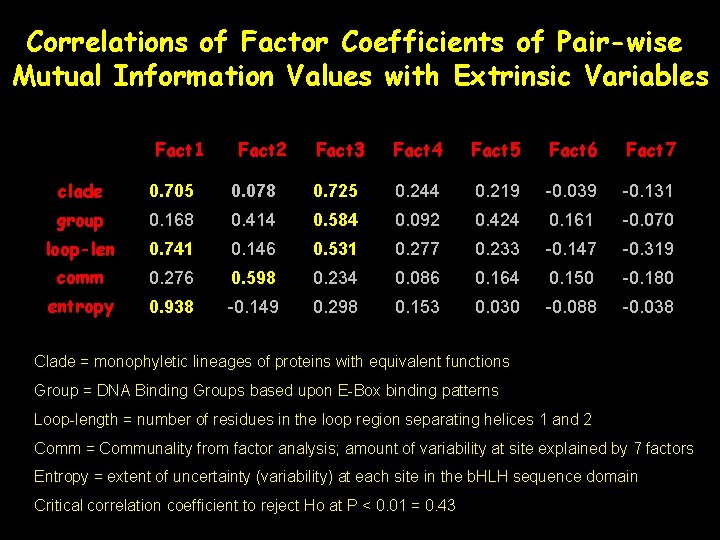
Correlations of Factor Coefficients of Pair-wise Mutual Information Values with Extrinsic Variables Fact 1 Fact 2 Fact 3 Fact 4 Fact 5 Fact 6 Fact 7 clade 0. 705 0. 078 0. 725 0. 244 0. 219 -0. 039 -0. 131 group 0. 168 0. 414 0. 584 0. 092 0. 424 0. 161 -0. 070 loop-len 0. 741 0. 146 0. 531 0. 277 0. 233 -0. 147 -0. 319 comm 0. 276 0. 598 0. 234 0. 086 0. 164 0. 150 -0. 180 entropy 0. 938 -0. 149 0. 298 0. 153 0. 030 -0. 088 -0. 038 Clade = monophyletic lineages of proteins with equivalent functions Group = DNA Binding Groups based upon E-Box binding patterns Loop-length = number of residues in the loop region separating helices 1 and 2 Comm = Communality from factor analysis; amount of variability at site explained by 7 factors Entropy = extent of uncertainty (variability) at each site in the b. HLH sequence domain Critical correlation coefficient to reject Ho at P < 0. 01 = 0. 43
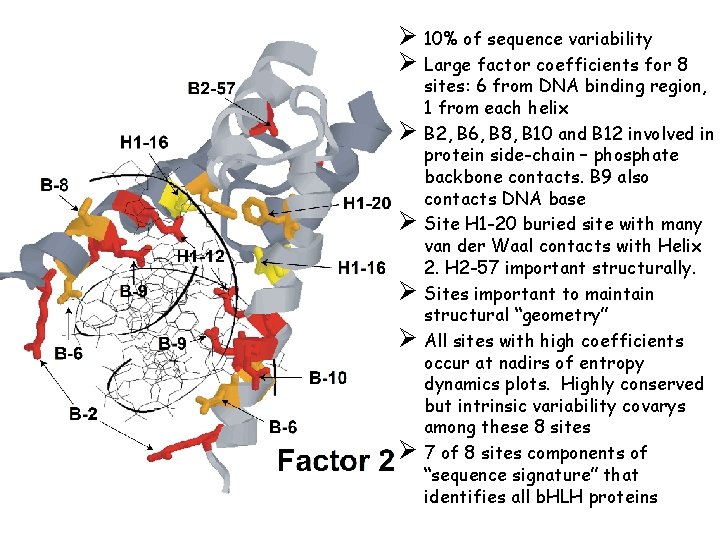
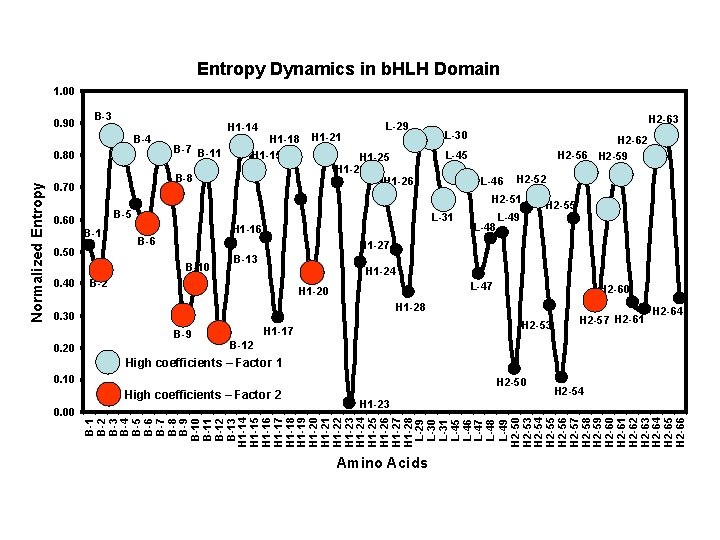
Ø 10% of sequence variability Ø Large factor coefficients for 8 Ø Ø Ø sites: 6 from DNA binding region, 1 from each helix B 2, B 6, B 8, B 10 and B 12 involved in protein side-chain – phosphate backbone contacts. B 9 also contacts DNA base Site H 1 -20 buried site with many van der Waal contacts with Helix 2. H 2 -57 important structurally. Sites important to maintain structural “geometry” All sites with high coefficients occur at nadirs of entropy dynamics plots. Highly conserved but intrinsic variability covarys among these 8 sites 7 of 8 sites components of “sequence signature” that identifies all b. HLH proteins
Entropy Dynamics in b. HLH Domain 1. 00 0. 90 B-3 H 1 -14 B-7 B-11 H 1 -15 H 1 -22 H 1 -26 B-8 0. 70 B-5 B-1 0. 50 H 2 -62 H 2 -56 H 2 -59 L-45 L-46 L-31 H 1 -16 B-6 H 2 -52 L-48 H 2 -55 L-49 H 1 -27 B-10 B-13 H 1 -24 B-2 L-47 H 1 -20 H 2 -60 H 1 -28 0. 30 B-9 0. 20 L-30 H 2 -51 0. 60 0. 40 H 1 -21 H 2 -63 H 2 -53 H 1 -17 H 2 -57 H 2 -61 H 2 -64 B-12 High coefficients – Factor 1 0. 10 Highincoefficients – Factor 2 Packed sites Max are underlined 0. 00 H 2 -54 H 1 -23 B-1 B-2 B-3 B-4 B-5 B-6 B-7 B-8 B-9 B-10 B-11 B-12 B-13 H 1 -14 H 1 -15 H 1 -16 H 1 -17 H 1 -18 H 1 -19 H 1 -20 H 1 -21 H 1 -22 H 1 -23 H 1 -24 H 1 -25 H 1 -26 H 1 -27 H 1 -28 L-29 L-30 L-31 L-45 L-46 L-47 L-48 L-49 H 2 -50 H 2 -53 H 2 -54 H 2 -55 H 2 -56 H 2 -57 H 2 -58 H 2 -59 H 2 -60 H 2 -61 H 2 -62 H 2 -63 H 2 -64 H 2 -65 H 2 -66 Normalized Entropy 0. 80 H 1 -18 L-29 Amino Acids
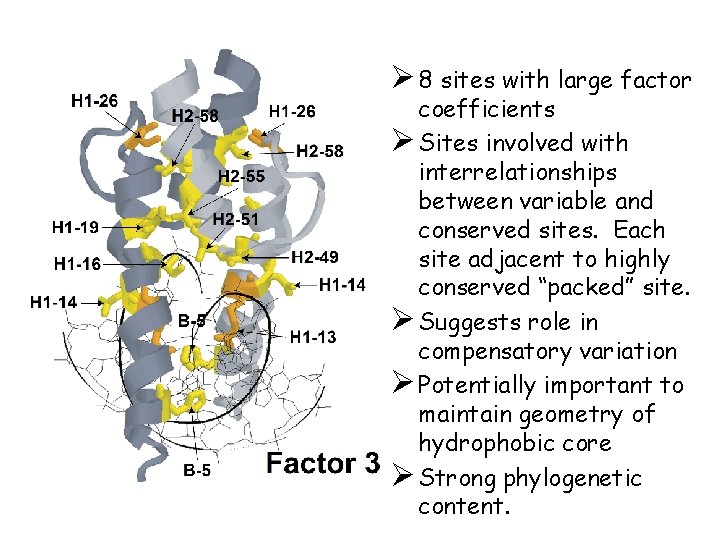
Ø 8 sites with large factor coefficients Ø Sites involved with interrelationships between variable and conserved sites. Each site adjacent to highly conserved “packed” site. Ø Suggests role in compensatory variation Ø Potentially important to maintain geometry of hydrophobic core Ø Strong phylogenetic content.
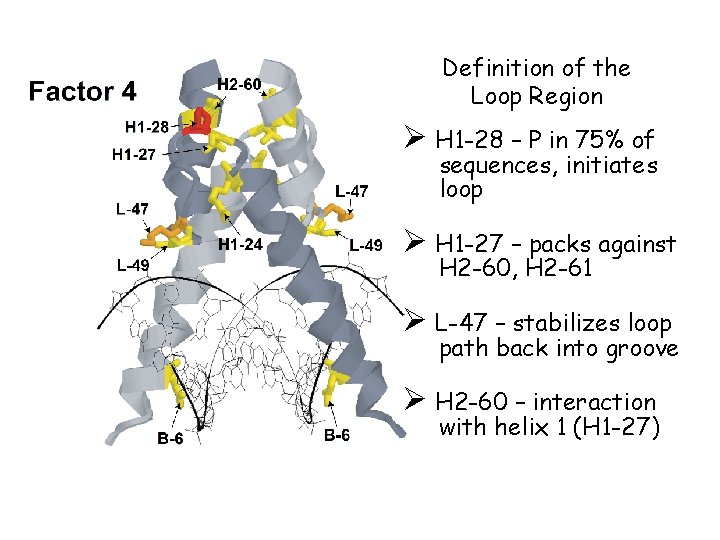
Definition of the Loop Region Ø H 1 -28 – P in 75% of sequences, initiates loop Ø H 1 -27 – packs against H 2 -60, H 2 -61 Ø L-47 – stabilizes loop path back into groove Ø H 2 -60 – interaction with helix 1 (H 1 -27)
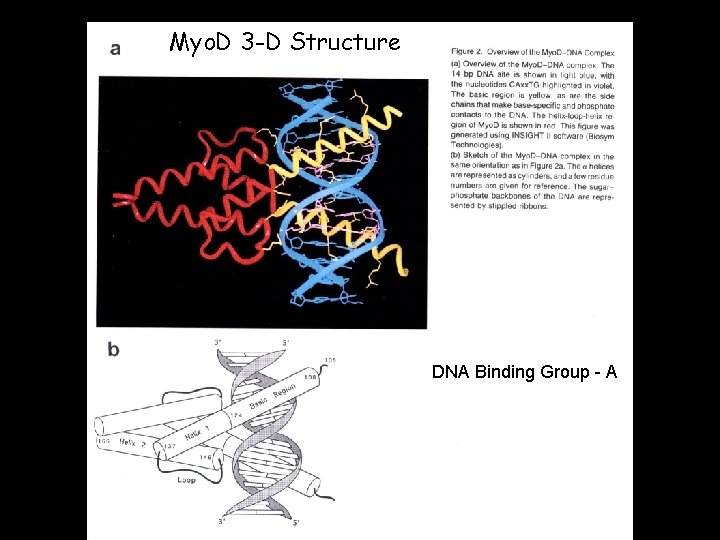
Myo. D 3 -D Structure DNA Binding Group - A
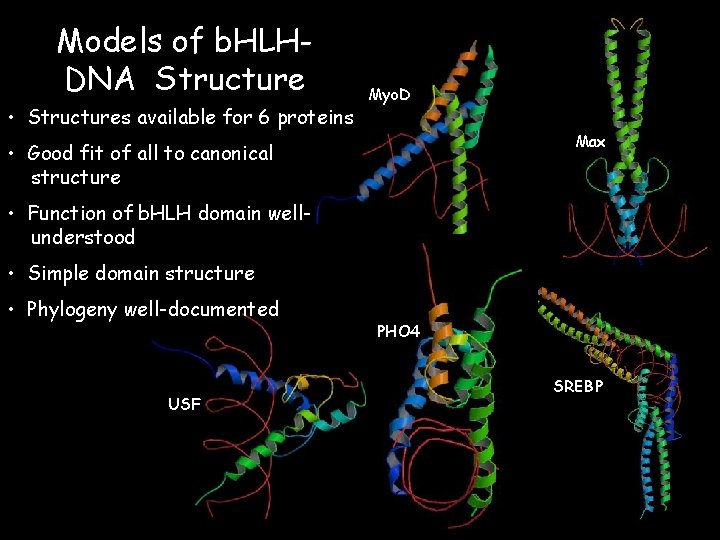
Models of b. HLHDNA Structure • Structures available for 6 proteins Myo. D Max • Good fit of all to canonical structure • Function of b. HLH domain wellunderstood • Simple domain structure • Phylogeny well-documented USF PHO 4 SREBP
- Slides: 34