Turner and Berg Turner and Berg Adaptation General

Turner and Berg

Turner and Berg

Adaptation
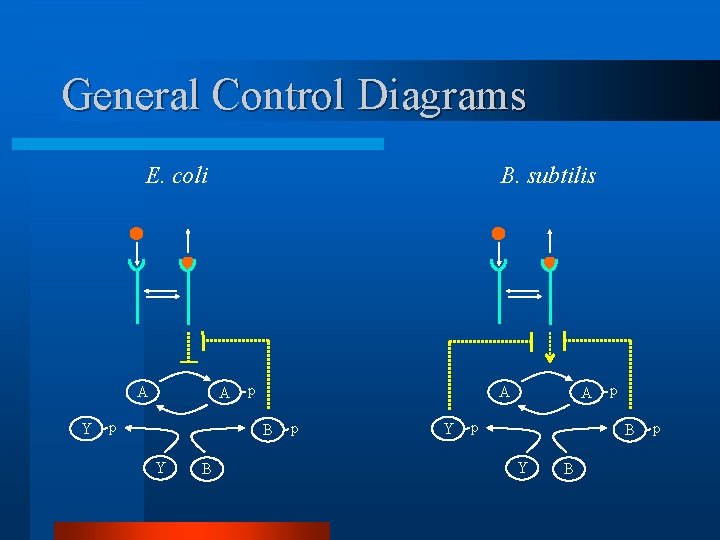
General Control Diagrams E. coli A Y B. subtilis A p p A B Y B p Y A p p B Y B p
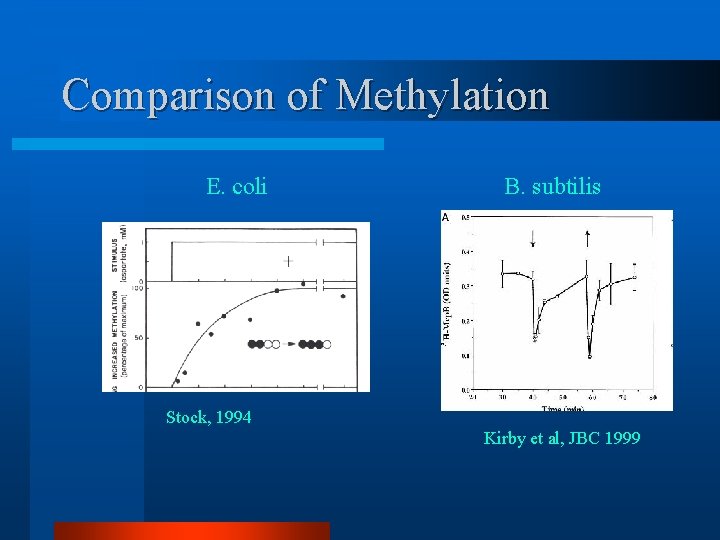
Comparison of Methylation E. coli B. subtilis Stock, 1994 Kirby et al, JBC 1999

A Simple Model of Adaptation Leibler Integral Feedback E. coli

Saturating Reaction Che. Bp Reaction Rate demethylation Che. R methylation Activity E. coli
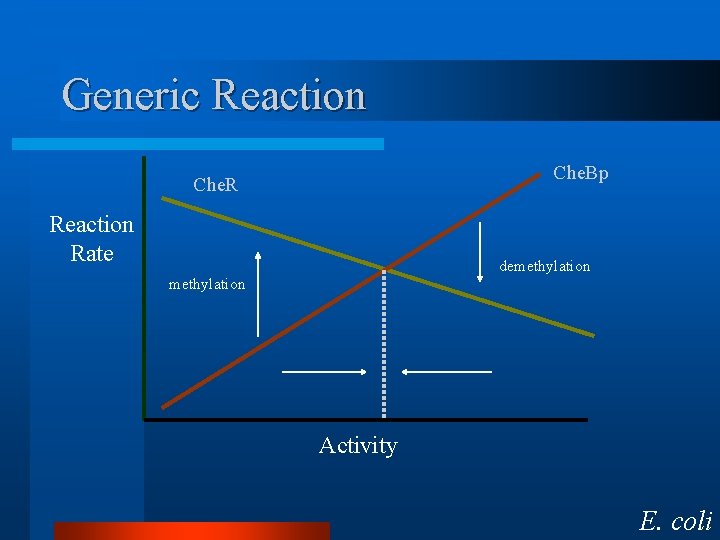
Generic Reaction Che. Bp Che. R Reaction Rate demethylation Activity E. coli

What is Inactivity? How do we create a mechanism for this? The challenge in B. subtilis E. coli

Activity Model Activity ligand no ligand methylation E. coli
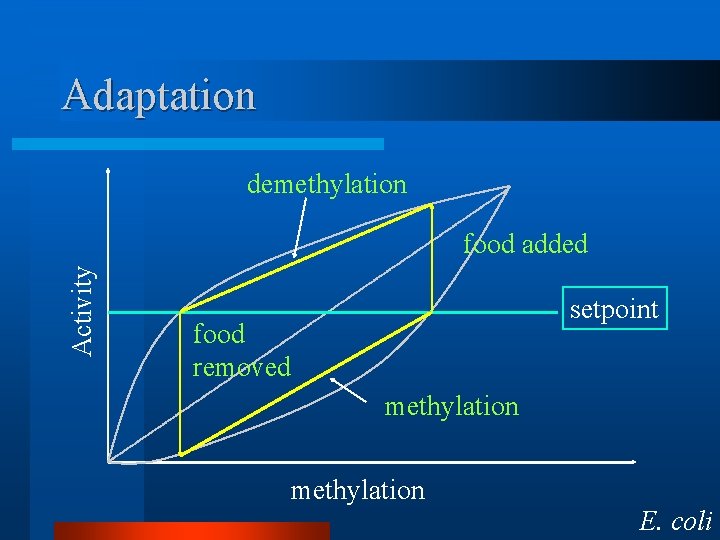
Adaptation demethylation Activity food added setpoint food removed methylation E. coli
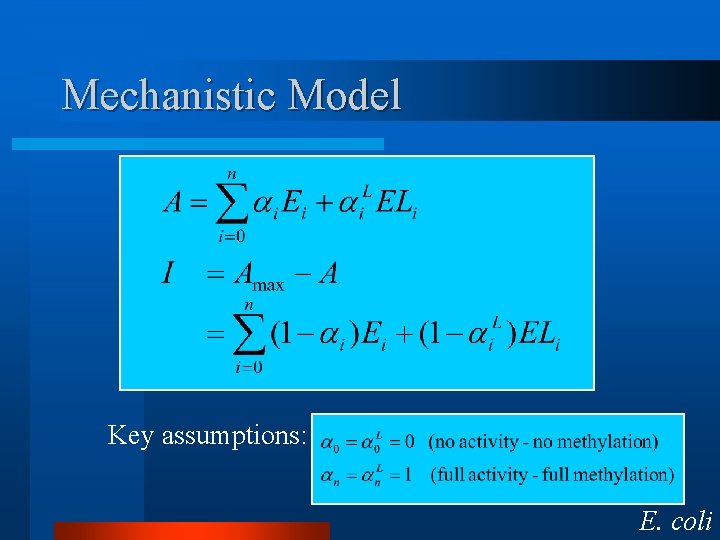
Mechanistic Model Key assumptions: E. coli

Why is are these assumption important? - Fully methylated receptors are not methylated. -Demethylated receptors are not demethylated. E. coli

So if we put this all together, we get robust adaptation. -Robustness so long as the previous conditions hold. -Can include enzyme kinetics, though a single enzyme controls each force. Che. R and Che. B. E. coli

Why is this a challenge for subtilis? l Receptor methylation increases and decreases activity. l Example of “Rein Control” l Unmethylated receptor has variable activity. l Four forces! – Methylation/demethylation site 1 – Methylation/demethylation site 2 B. subtilis
Rein Control Right Left B. subtilis

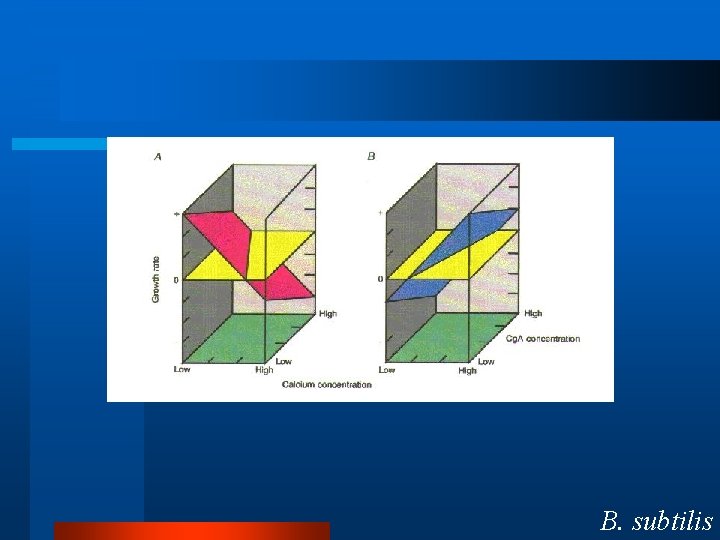
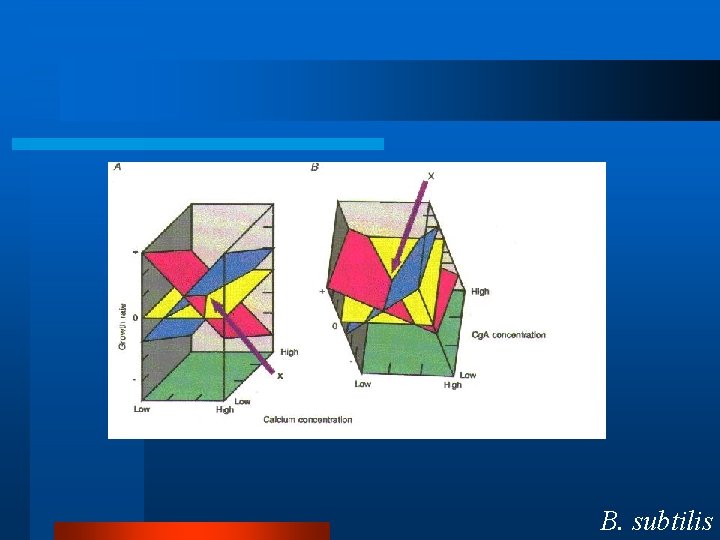
Control Motif l Physiology – Calcium and Glucose Homeostasis. l Two forces – One Increases – One Decreases l Two feedback loops. B. subtilis
B. subtilis
B. subtilis

B. subtilis appears to use this control motif. So, we have two integral feedback loops. -One increases activity. -One decreases activity. What is the molecular mechanism? B. subtilis

Activity Model Site 1 Activity Ligand No Ligand Che. Yp Feedback Site 2 B. subtilis
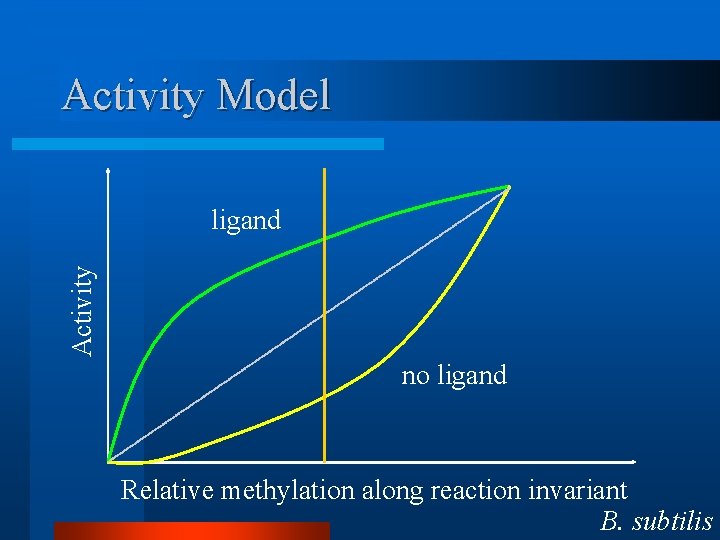
Activity Model Activity ligand no ligand Relative methylation along reaction invariant B. subtilis
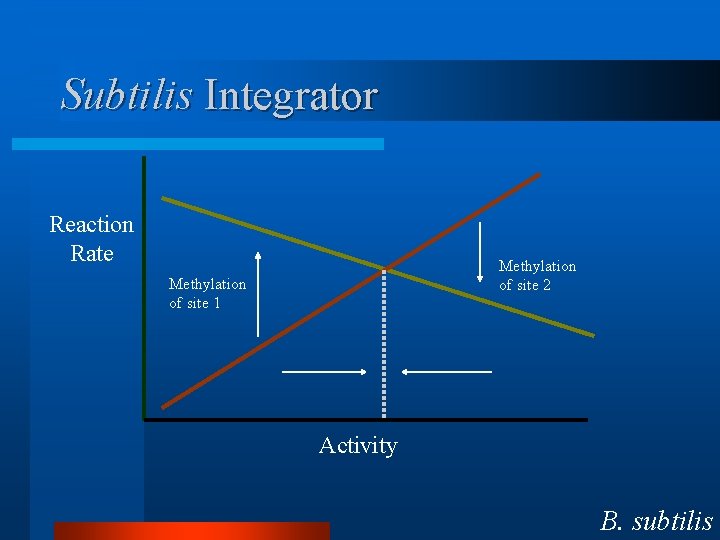
Subtilis Integrator Reaction Rate Methylation of site 2 Methylation of site 1 Activity B. subtilis
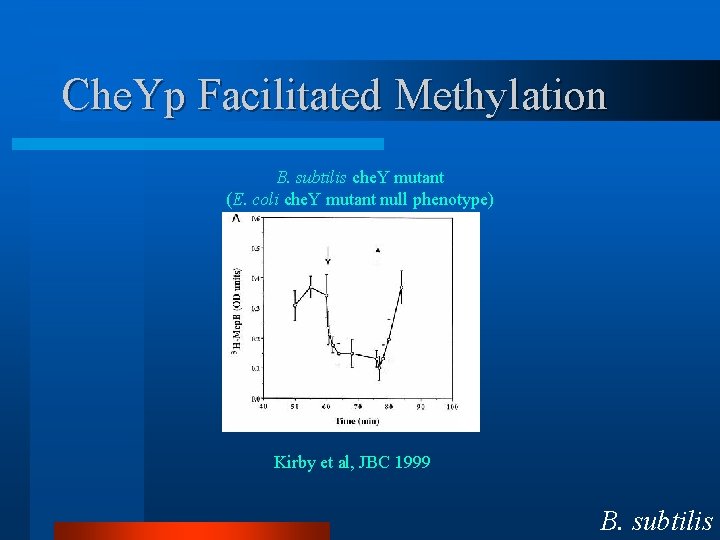
Che. Yp Facilitated Methylation B. subtilis che. Y mutant (E. coli che. Y mutant null phenotype) Kirby et al, JBC 1999 B. subtilis
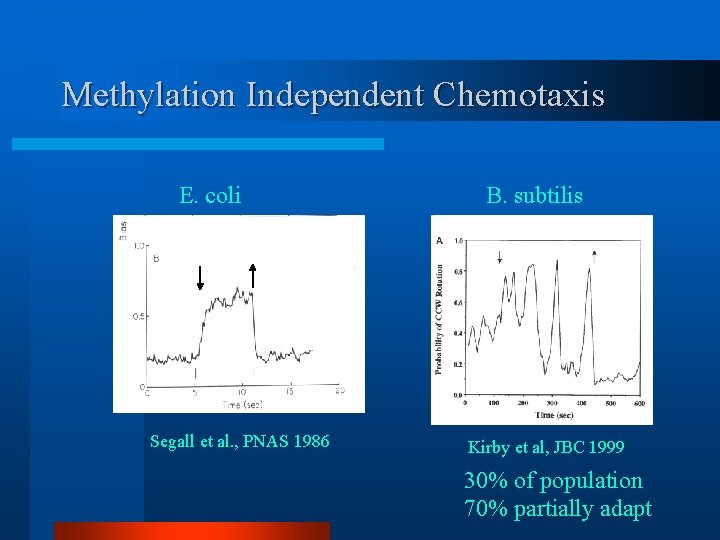
Methylation Independent Chemotaxis E. coli Segall et al. , PNAS 1986 B. subtilis Kirby et al, JBC 1999 30% of population 70% partially adapt
- Slides: 25