Tree Reconstruction Phylogenetic tree Nodes DNA RNA mt







- Slides: 13

Tree Reconstruction

Phylogenetic tree • Nodes – DNA (RNA, mt. DNA) sequences, proteins, species = taxonomic units (TUs) • Branches – ancestral relations between Tus • Terminal (extant) nodes, leaves – OTUs (O for operational)

Tree reconstruction • Neighbor joining (Distance) methods • Maximum parsimony methods (W. Fitch) • Maximum likelihood methods (J. Felsenstein) W. H. Li, “Molecular Evolution”, 1997

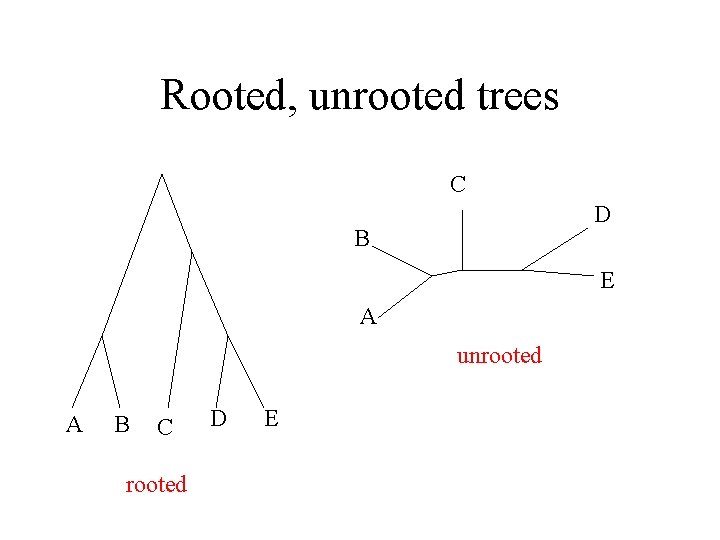
Rooted, unrooted trees C D B E A unrooted A B C rooted D E

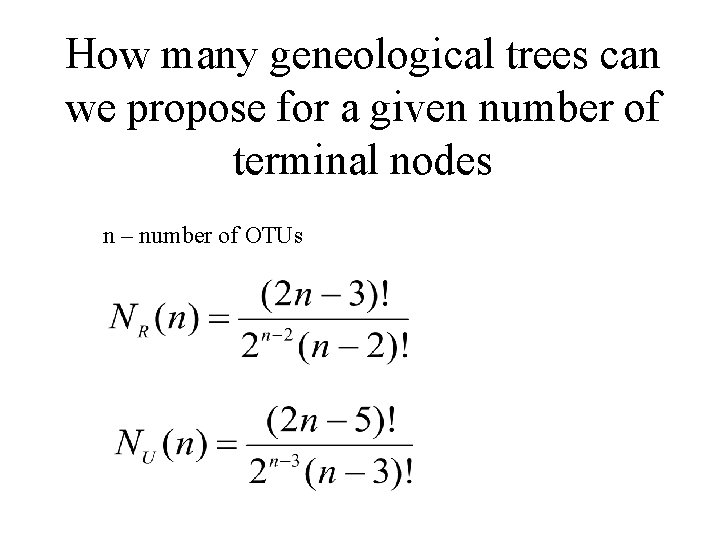
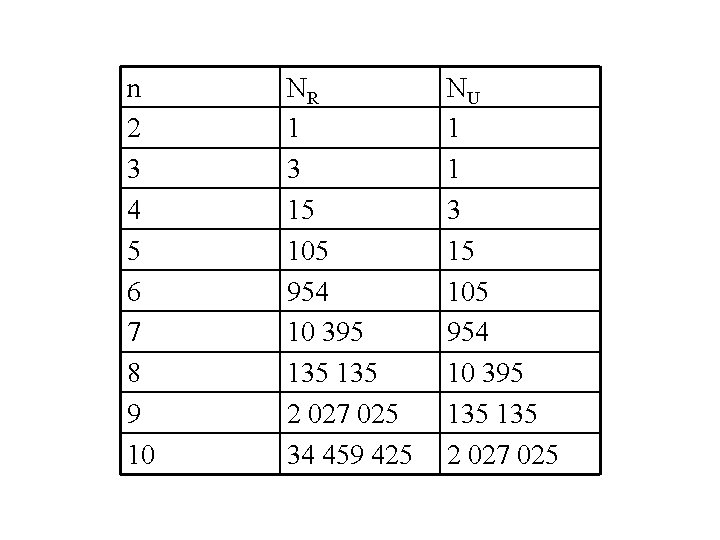
How many geneological trees can we propose for a given number of terminal nodes n – number of OTUs

n 2 3 4 5 6 7 8 9 10 NR 1 3 15 105 954 10 395 135 2 027 025 34 459 425 NU 1 1 3 15 105 954 10 395 135 2 027 025

Neighbor joining • UPGMA – unweighted pair group method with arithmetic mean • • Start from distance matrix (*) Find the minimum distance OTUs And merge them Update distance matrix, go to (*)

Maximum parsimony • Principle of max parsimony searches for a tree that requires the smallest number of evolutionary changes to explain differences among OTUs • Informative sites

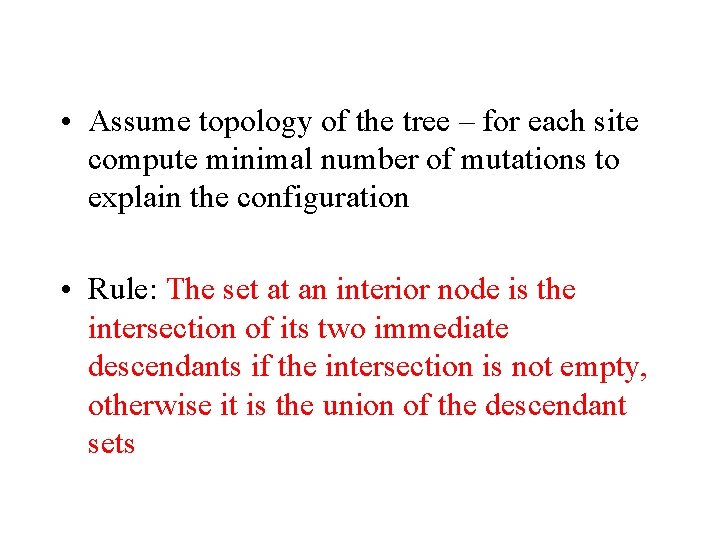
• Assume topology of the tree – for each site compute minimal number of mutations to explain the configuration • Rule: The set at an interior node is the intersection of its two immediate descendants if the intersection is not empty, otherwise it is the union of the descendant sets

• The index of the tree is the sum of indices for all informative sites • Go through all possible trees to search for optimal one

Maximum likelihood • Need a probabilistic model for nucleotide substitution A, C, T, G – 1, 2, 3, 4 time=0 We analyze evoution of one site S. Given S=i, time=0 what is the probability of S=j, time=t

• Compute the likelihood function for a given tree • Go through all possible trees to search for optimal one

Software PHYLIP (Phylogeny Inference Package) Version 3. 57 c by Joseph Felsenstein July, 1995