Translation SBI 4 U 1 Components of Translation
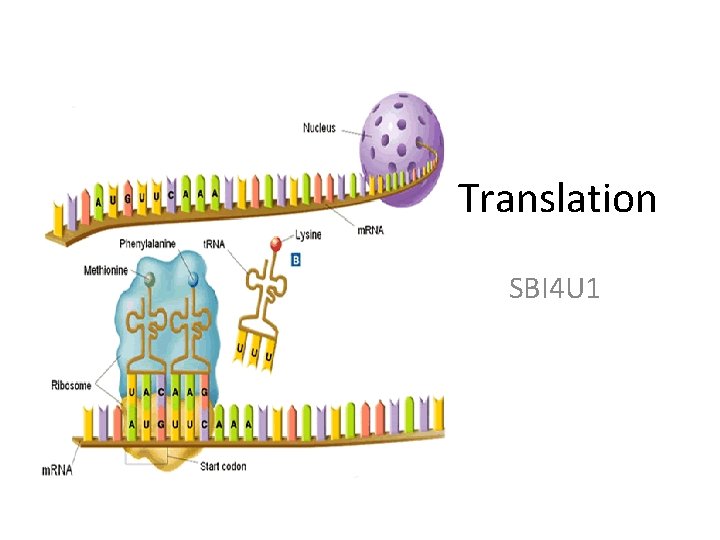
Translation SBI 4 U 1
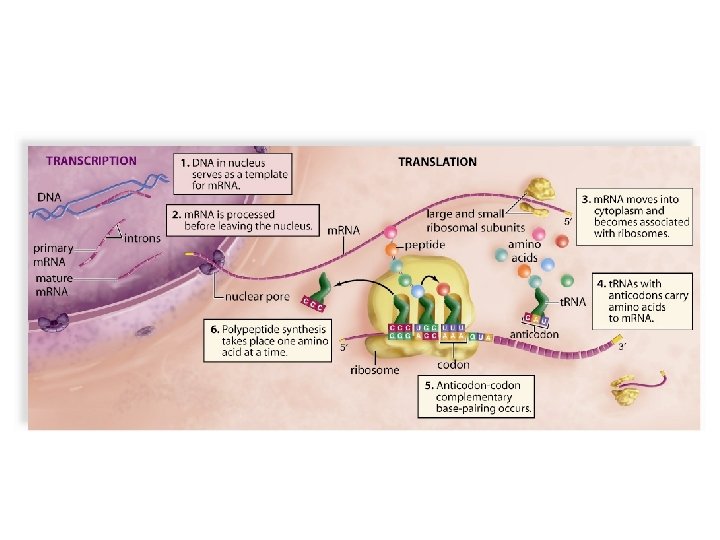
Components of Translation • m. RNA – Determines amino acid sequence • t. RNA – Contains anticodon that pairs w/ a codon on m. RNA w/ amino acid attached • Ribosomes – Made of r. RNA and proteins (protein synthesis) • Translation factors – Proteins needed for translation
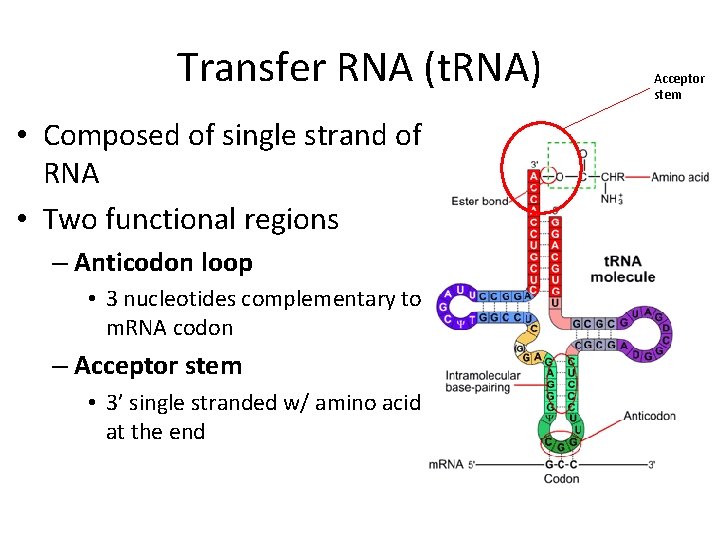
Transfer RNA (t. RNA) • Composed of single strand of RNA • Two functional regions – Anticodon loop • 3 nucleotides complementary to m. RNA codon – Acceptor stem • 3’ single stranded w/ amino acid at the end Acceptor stem
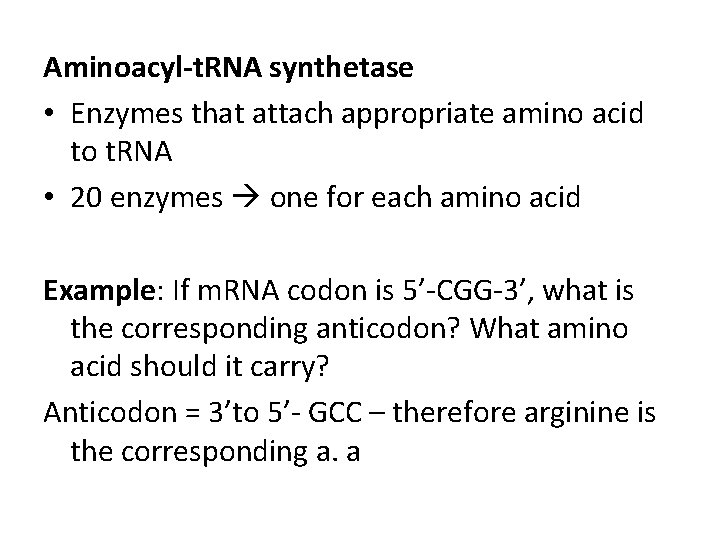
Aminoacyl-t. RNA synthetase • Enzymes that attach appropriate amino acid to t. RNA • 20 enzymes one for each amino acid Example: If m. RNA codon is 5’-CGG-3’, what is the corresponding anticodon? What amino acid should it carry? Anticodon = 3’to 5’- GCC – therefore arginine is the corresponding a. a
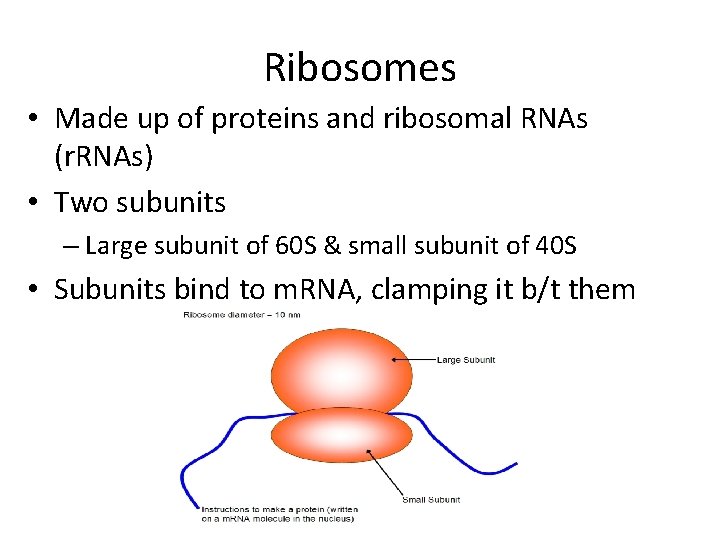
Ribosomes • Made up of proteins and ribosomal RNAs (r. RNAs) • Two subunits – Large subunit of 60 S & small subunit of 40 S • Subunits bind to m. RNA, clamping it b/t them
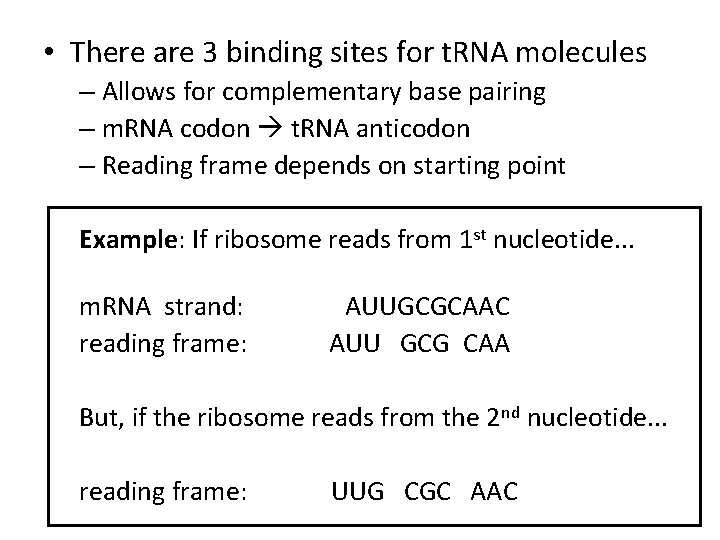
• There are 3 binding sites for t. RNA molecules – Allows for complementary base pairing – m. RNA codon t. RNA anticodon – Reading frame depends on starting point Example: If ribosome reads from 1 st nucleotide. . . m. RNA strand: reading frame: AUUGCGCAAC AUU GCG CAA But, if the ribosome reads from the 2 nd nucleotide. . . reading frame: UUG CGC AAC
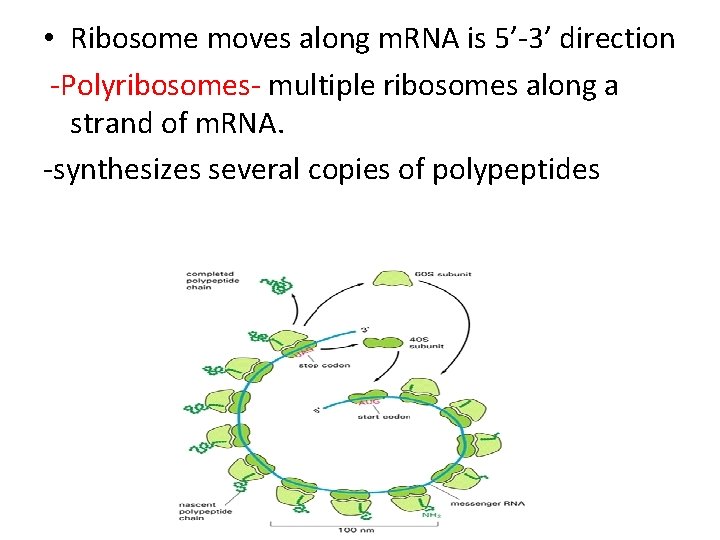
• Ribosome moves along m. RNA is 5’-3’ direction -Polyribosomes- multiple ribosomes along a strand of m. RNA. -synthesizes several copies of polypeptides

3 Stages in Translation: 1. Initiation 2. Elongation 3. Termination Note: these are NOT similar to DNA replication and transcription
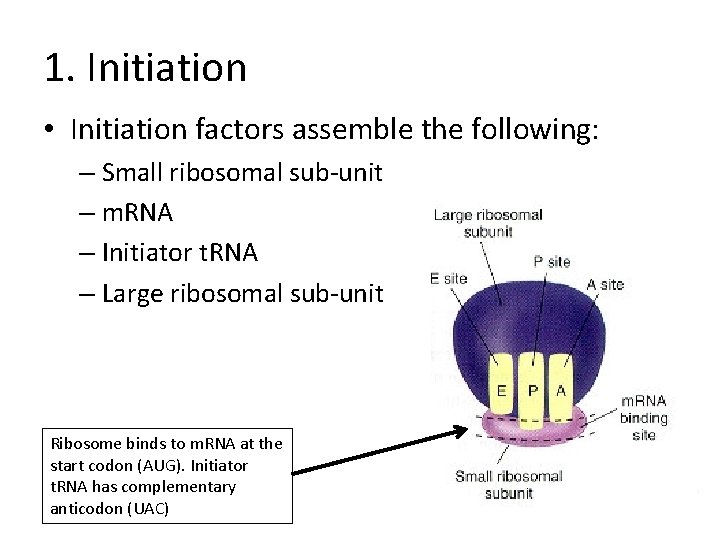
1. Initiation • Initiation factors assemble the following: – Small ribosomal sub-unit – m. RNA – Initiator t. RNA – Large ribosomal sub-unit Ribosome binds to m. RNA at the start codon (AUG). Initiator t. RNA has complementary anticodon (UAC)
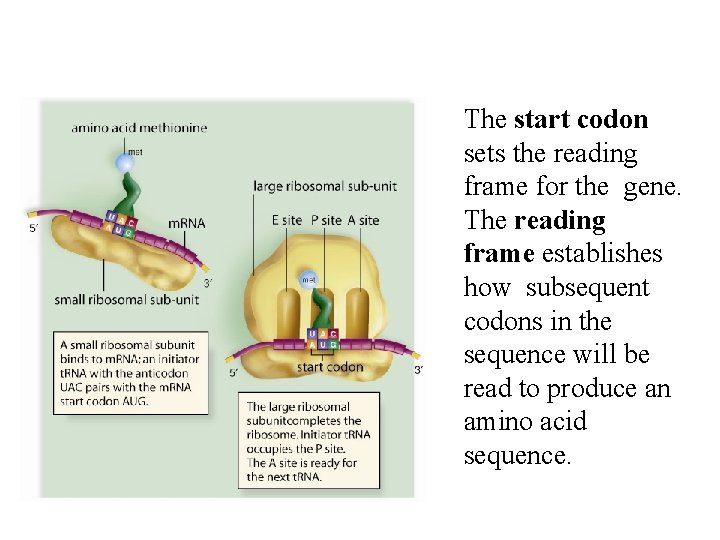
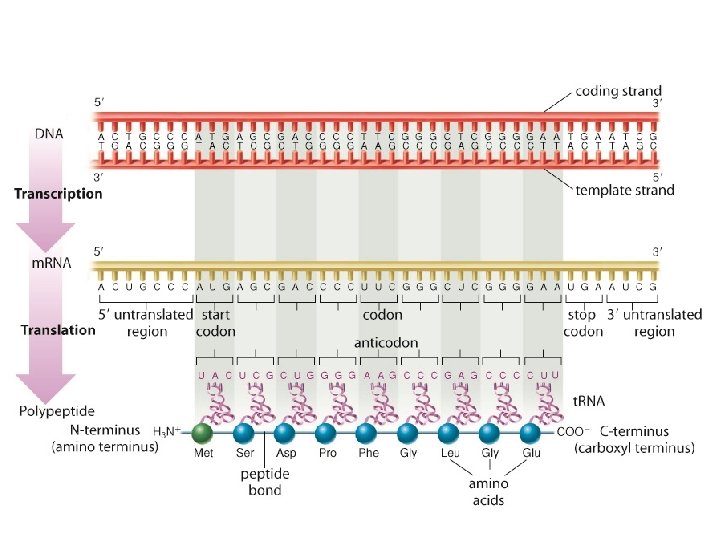
The start codon sets the reading frame for the gene. The reading frame establishes how subsequent codons in the sequence will be read to produce an amino acid sequence.
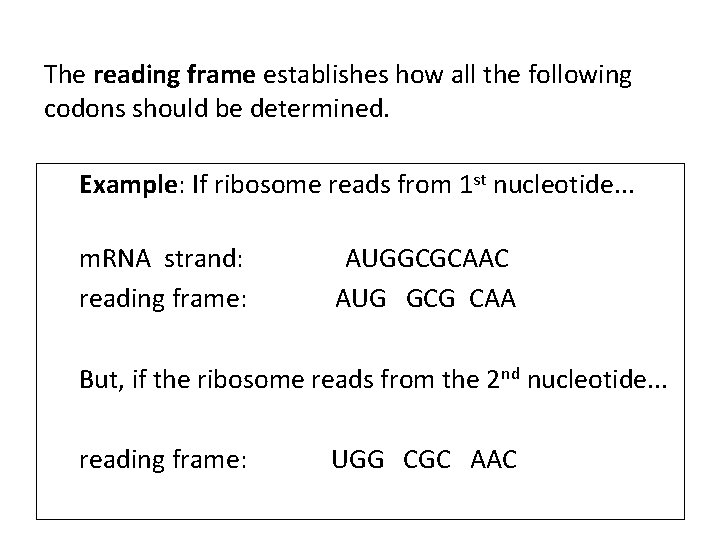
The reading frame establishes how all the following codons should be determined. Example: If ribosome reads from 1 st nucleotide. . . m. RNA strand: reading frame: AUGGCGCAAC AUG GCG CAA But, if the ribosome reads from the 2 nd nucleotide. . . reading frame: UGG CGC AAC
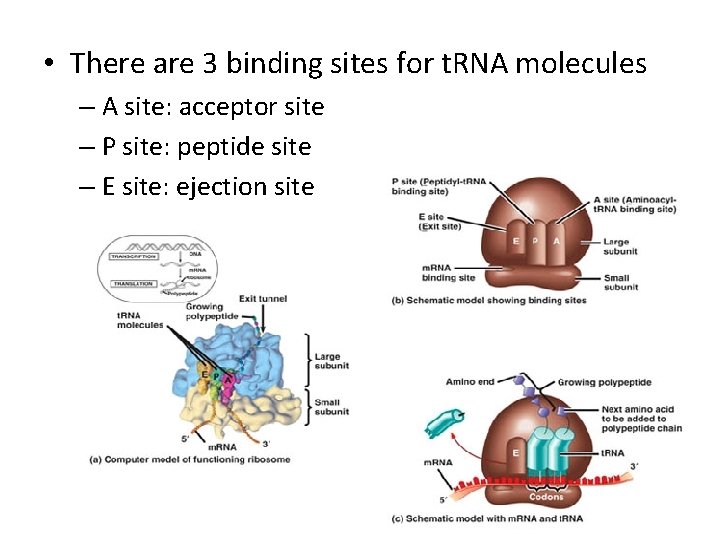
• There are 3 binding sites for t. RNA molecules – A site: acceptor site – P site: peptide site – E site: ejection site
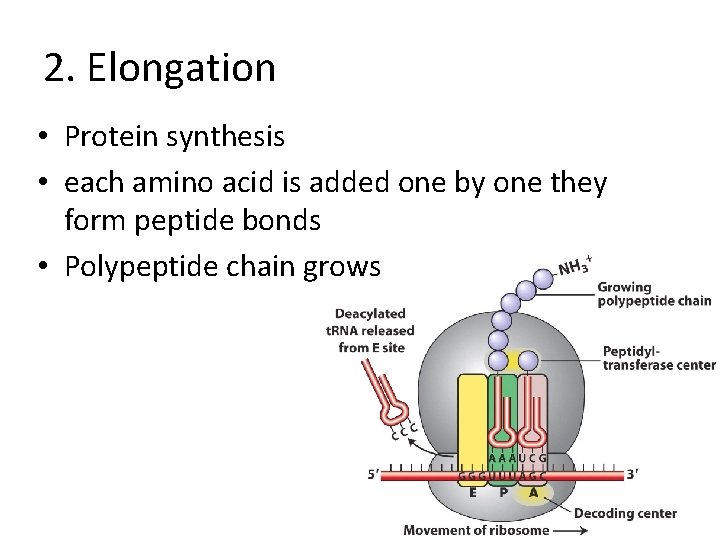
2. Elongation • Protein synthesis • each amino acid is added one by one they form peptide bonds • Polypeptide chain grows
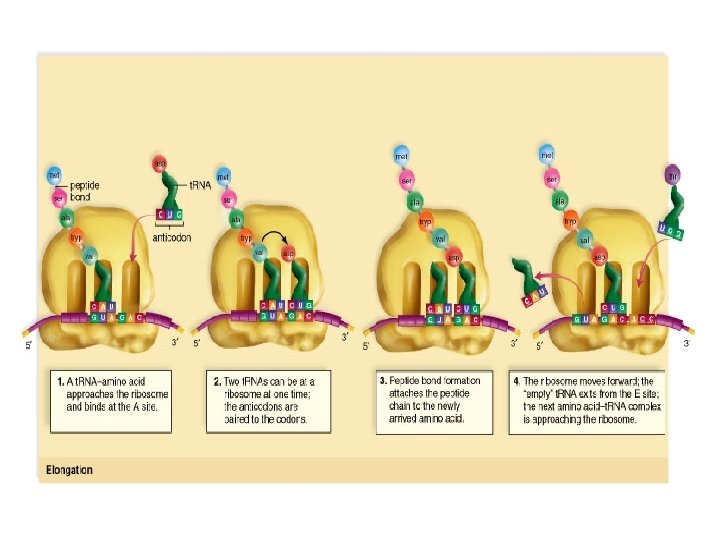

3. Termination • Stop codon on m. RNA is reached • Release factor cleaves polypeptide • Protein is modified, folded and sent to appropriate area of the cell
Translation Animation http: //www. stolaf. edu/people/giannini/flashanima t/molgenetics/translation. swf For Prokaryotes (similar): https: //highered. mcgrawhill. com/sites/0072507470/student_view 0/chapter 3/animation__how_translation_work s. html
DNA Mutations and Effects of Mutagens Mutation: permanent change in nucleotide sequence in DNA • Mutations are copied during DNA replication • Mutations in reproductive cells are passed on to future generations • Mutations in somatic cells don’t effect future generations
Mutations are grouped into 2 categories: 1) Single Gene Mutations -changes in nucleotide sequence -involves 1 gene 2) Chromosomal Mutations -changes in chromosomes -involves many genes
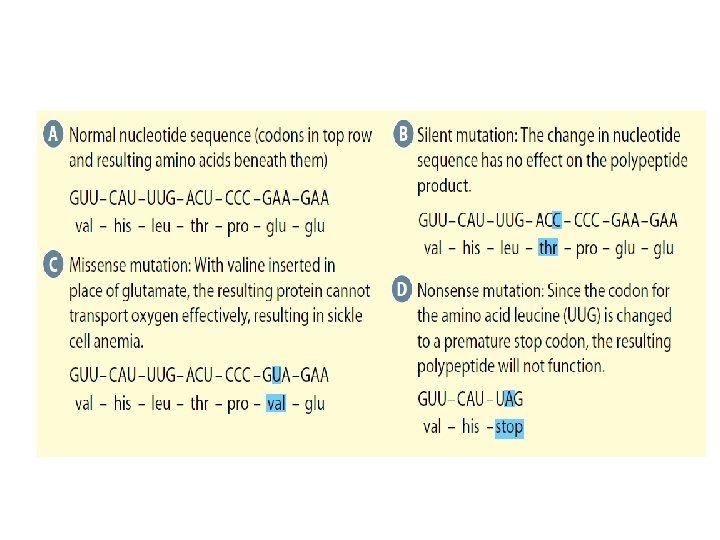
(1)Single Gene Mutations: Silent Mutation: no change in amino acid sequence Missense Mutation: changes amino acid sequence and can be harmful Nonesense Mutation: converts codon for amino acid to a stop codon
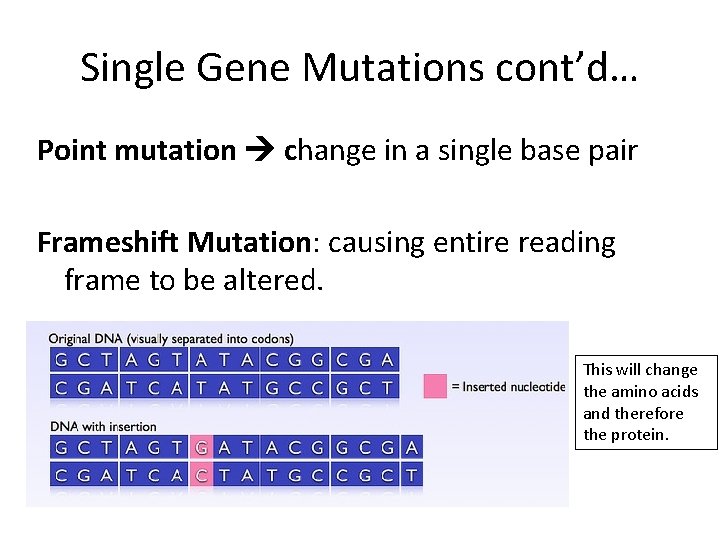
Single Gene Mutations cont’d… Point mutation change in a single base pair Frameshift Mutation: causing entire reading frame to be altered. This will change the amino acids and therefore the protein.
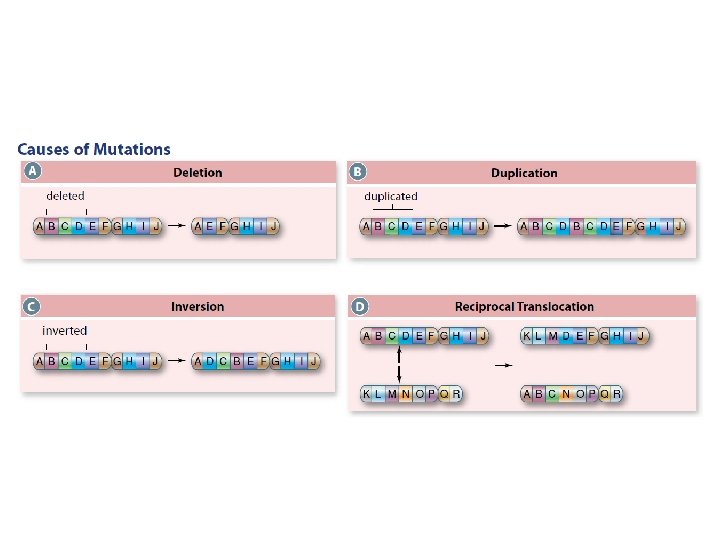
(2)Chromosome Mutations Rearrangement of genetic material on chromosome structure • deletion or duplication of portions of chromosomes • inversions (a chromosome segment is broken and re-inserted in the opposite direction) • translocations (a section of one chromosome is broken and fused to another chromosome
Causes of Mutations can be either spontaneous or induced. Spontaneous mutations may be caused by: a) incorrect base pairing by DNA polymerase during replication b) “transposition”; in which specific DNA sequences (called transposons) move within and between chromosomes
Induced Mutagens Induced mutations may be caused by: • Physical mutagens – X-rays tear through DNA and change structure – UV radiation distorts DNA molecule • Chemical mutagens – Molecule that enters nucleus and reacts w/ DNA – Nitrites (cured meats), gasoline fumes, cigarettes

DNA Repair • Recall: DNA polymerase “proof reads” as a mismatch repair mechanism • Specific and non-specific repair mechanisms also fix mutations
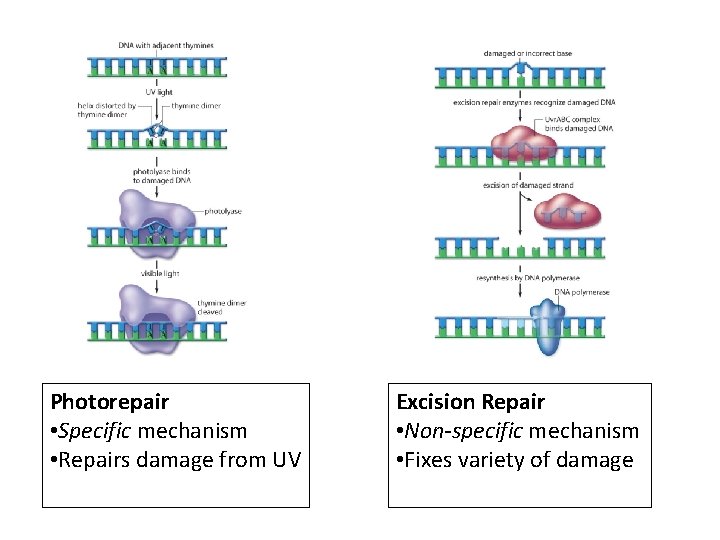
Photorepair • Specific mechanism • Repairs damage from UV Excision Repair • Non-specific mechanism • Fixes variety of damage

Learning Expectations. . . • Components of translation (structure/functions) – m. RNA – t. RNA – Ribosomes • • 3 stages in translation (in detail) Be able to go from DNA m. RNA A. A. sequence All single gene and chromosomal mutations Example of mutagens
- Slides: 30