Translation Gene Expression Regulators Transcriptional factors Chromatin structure





- Slides: 32

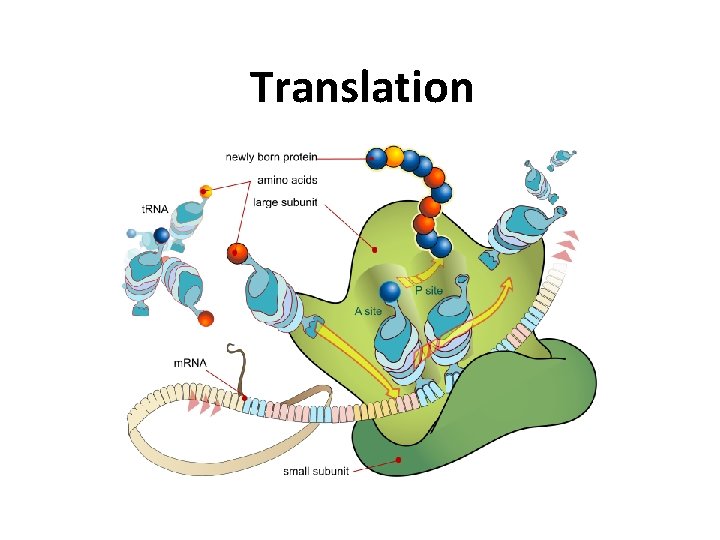
Translation

Gene Expression Regulators • • • Transcriptional factors Chromatin structure Non-coding RNA Translation initiation factors Efficiency of translation

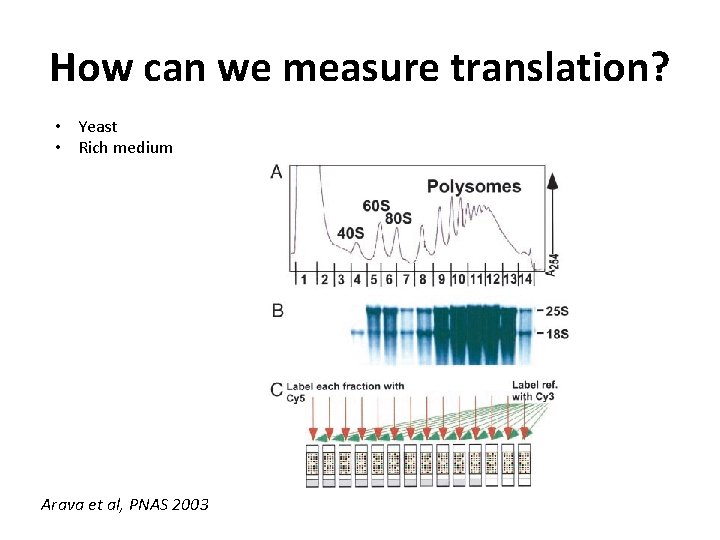
How can we measure translation? • Yeast • Rich medium Arava et al, PNAS 2003

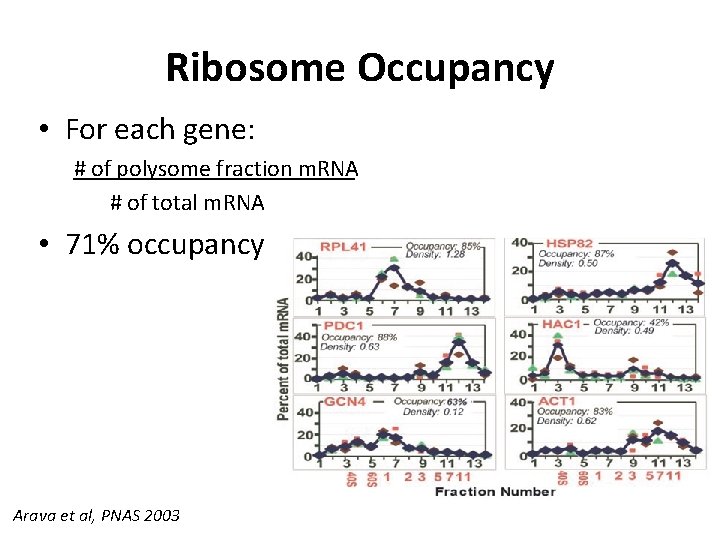
Ribosome Occupancy • For each gene: # of polysome fraction m. RNA # of total m. RNA • 71% occupancy Arava et al, PNAS 2003

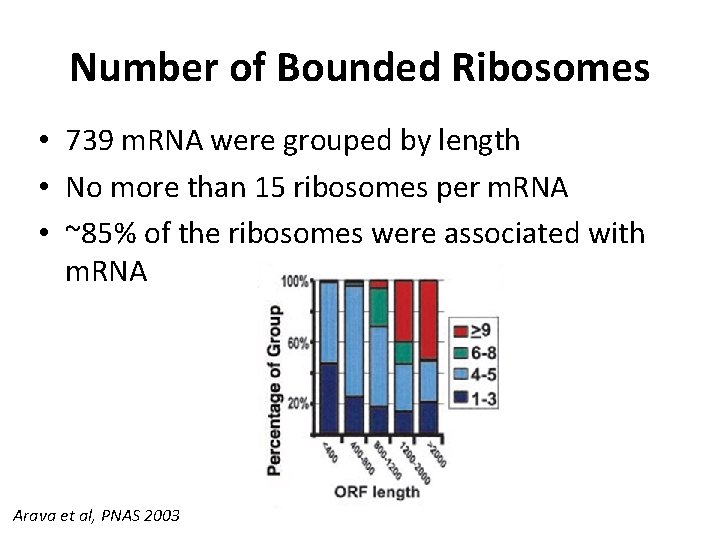
Number of Bounded Ribosomes • 739 m. RNA were grouped by length • No more than 15 ribosomes per m. RNA • ~85% of the ribosomes were associated with m. RNA Arava et al, PNAS 2003

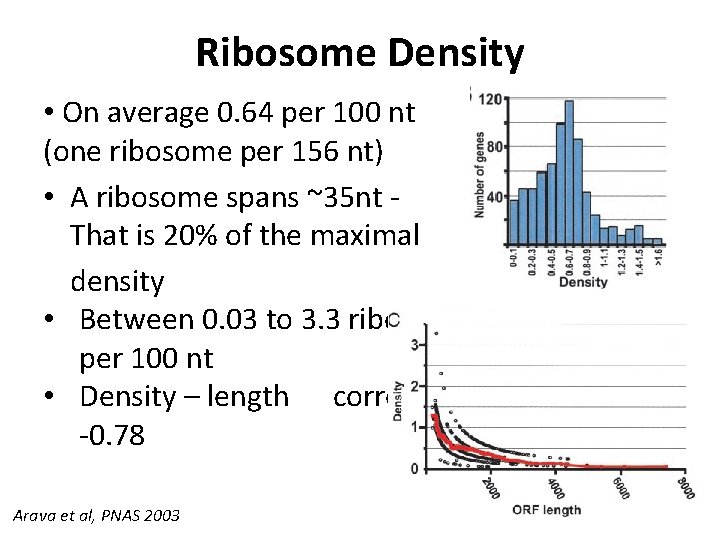
Ribosome Density • On average 0. 64 per 100 nt (one ribosome per 156 nt) • A ribosome spans ~35 nt That is 20% of the maximal density • Between 0. 03 to 3. 3 ribosomes per 100 nt • Density – length correlation: -0. 78 Arava et al, PNAS 2003

This is good, but limited. . • • Does not consider the ribosome position Only one state was analyzed Ignores m. RNA structure Was done only with one specie

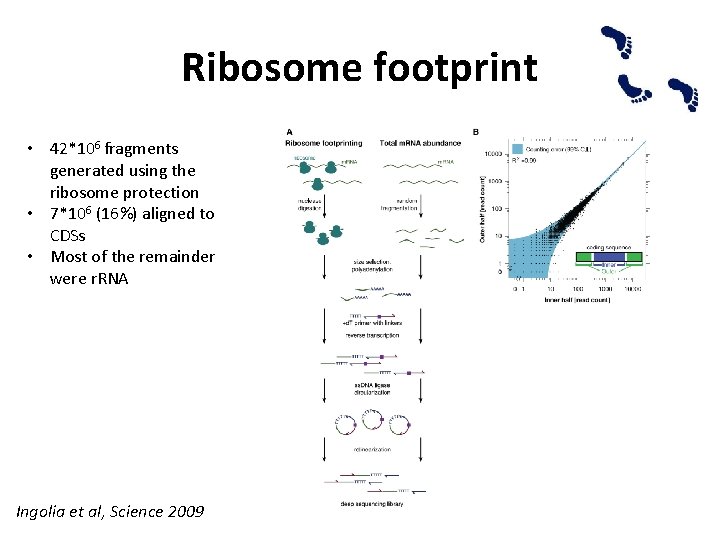
Ribosome footprint • Ribosome-profiling based on deep sequencing of ribosome-protected fragment – Robustly generate ribosome protected m. RNA fragments – Converts these RNA footprints into a library of DNA ready for sequencing – Measures the abundance of different footprints in this library

Ribosome footprint • 42*106 fragments generated using the ribosome protection • 7*106 (16%) aligned to CDSs • Most of the remainder were r. RNA Ingolia et al, Science 2009

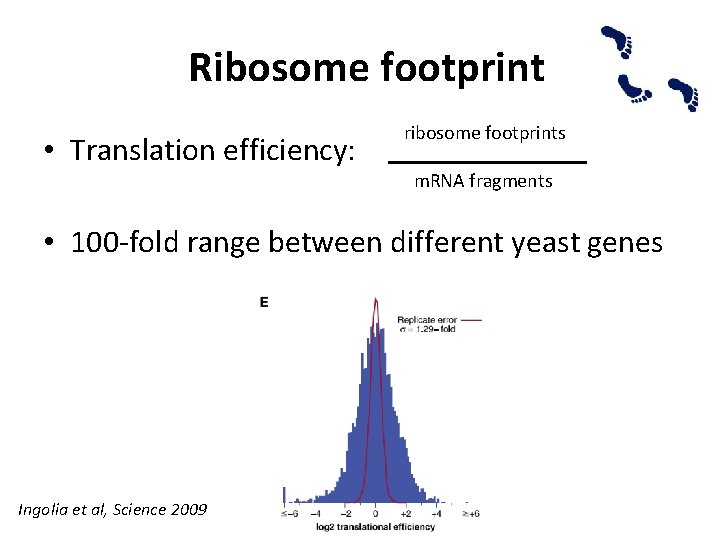
Ribosome footprint • Translation efficiency: ribosome footprints m. RNA fragments • 100 -fold range between different yeast genes Ingolia et al, Science 2009

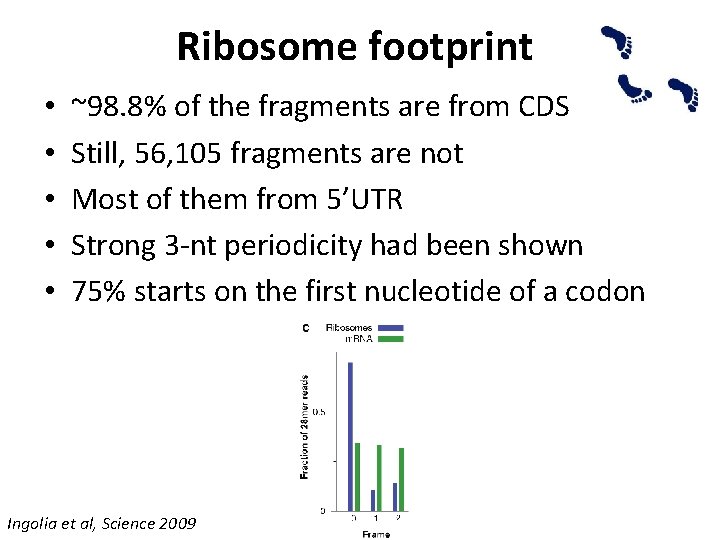
Ribosome footprint • • • ~98. 8% of the fragments are from CDS Still, 56, 105 fragments are not Most of them from 5’UTR Strong 3 -nt periodicity had been shown 75% starts on the first nucleotide of a codon Ingolia et al, Science 2009

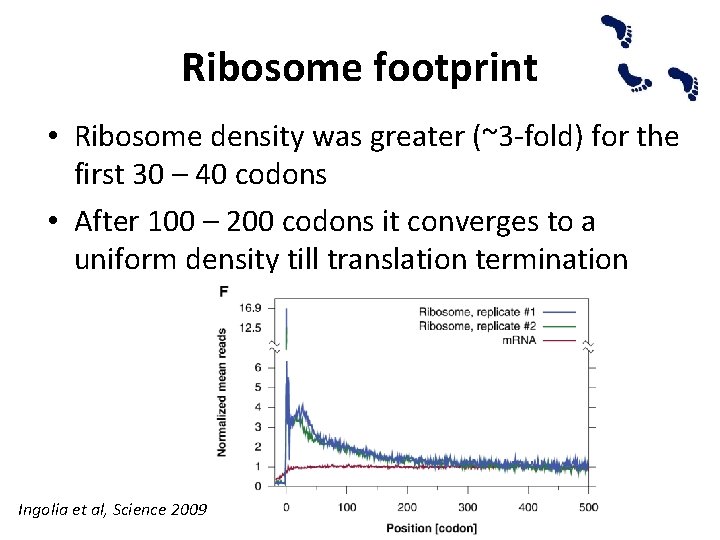
Ribosome footprint • Ribosome density was greater (~3 -fold) for the first 30 – 40 codons • After 100 – 200 codons it converges to a uniform density till translation termination Ingolia et al, Science 2009

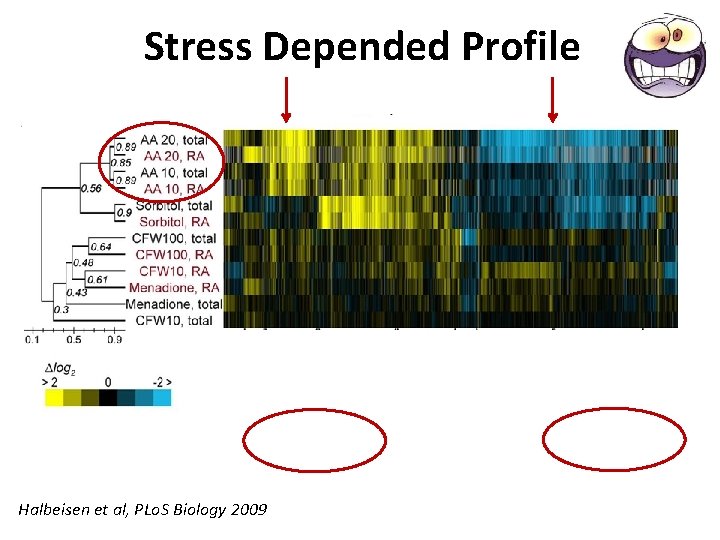
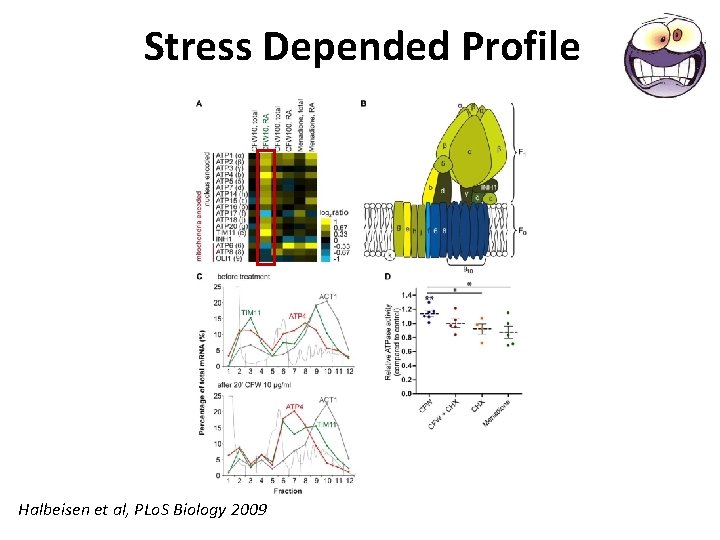
Stress Depended Profile • Severe stress – Environmental Stress Response (ESR) – A. A starvation, osmotic shock • Mild stress - stimuli that affect cellular homeostasis without triggering ESR – CFW, a cell-wall–perturbing agent – mild oxidative stress

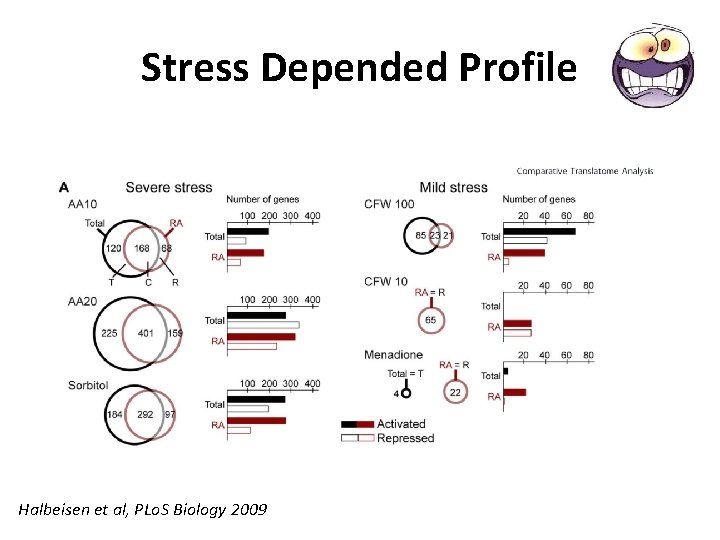
Stress Depended Profile Halbeisen et al, PLo. S Biology 2009

Stress Depended Profile Halbeisen et al, PLo. S Biology 2009

Stress Depended Profile Halbeisen et al, PLo. S Biology 2009

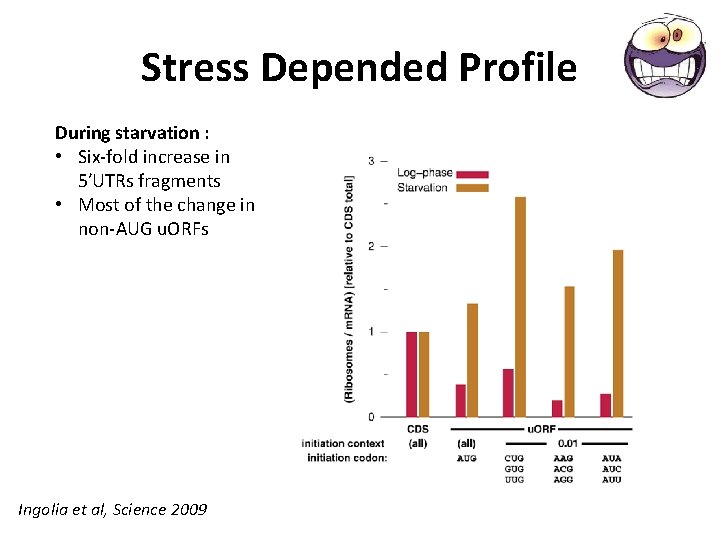
Stress Depended Profile During starvation : • Six-fold increase in 5′UTRs fragments • Most of the change in non-AUG u. ORFs Ingolia et al, Science 2009

So far. . • • • Ribosome occupancy per gene Number of ribosomes per m. RNA Ribosome density Ribosome location Metabolic activity can be regulated without the need of transcription

Quantification of Translation Efficiency • What is the optimal level of expression of a given protein ? – Protein translation is source consuming: • • • Ribosomes t. RNAs aminoacyl t. RNA synthetases Amino acids Translation factors Energy

Quantification of Translation Efficiency • CAI – Codon Adaptation Index: codon’s frequency in highly expressed genes frequency of the most abundant codon for that amino acid • t. AI – t. RNA Adaptation Index – Considering the t. RNA pool – Considering wobble rules • CAI/t. AI of an entire gene is computed as the geometric mean of the value of each codon

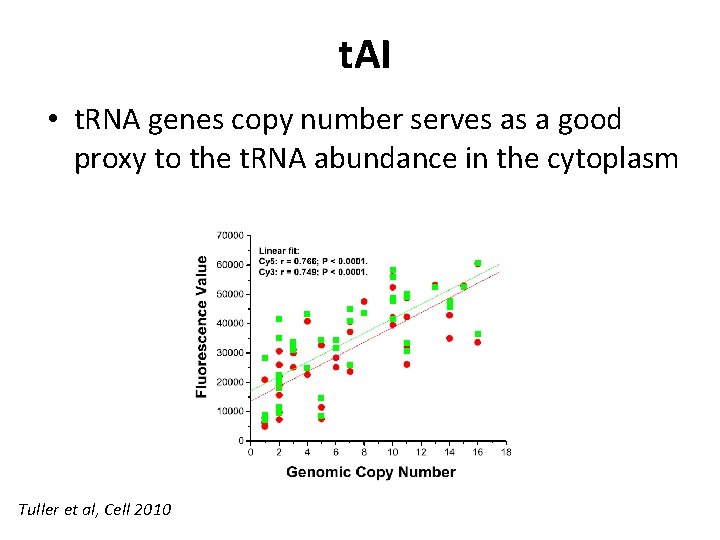
t. AI • t. RNA genes copy number serves as a good proxy to the t. RNA abundance in the cytoplasm Tuller et al, Cell 2010

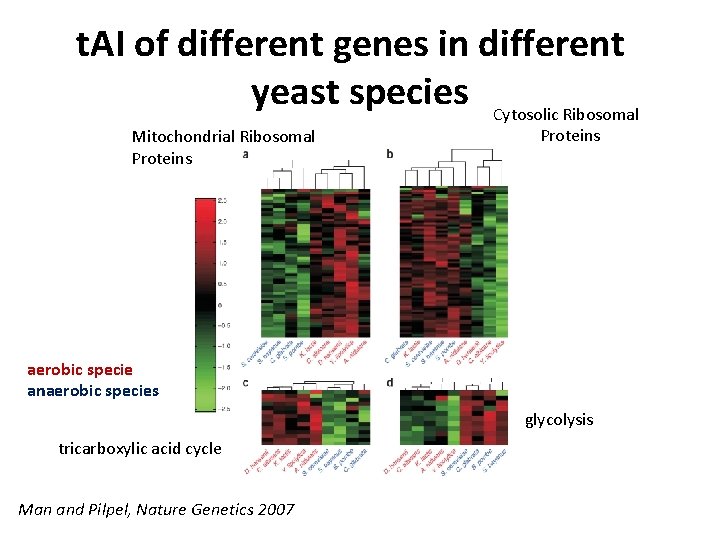
t. AI of different genes in different yeast species Cytosolic Ribosomal Mitochondrial Ribosomal Proteins aerobic specie anaerobic species glycolysis tricarboxylic acid cycle Man and Pilpel, Nature Genetics 2007

More factors to consider. . • Local pools of required t. RNAs might promote translation elongation efficiency • The order of the codons should be considered • Estimate the concentration of amino acidloaded t. RNAs • The t. RNA pool can vary in different tissue or different environmental conditions

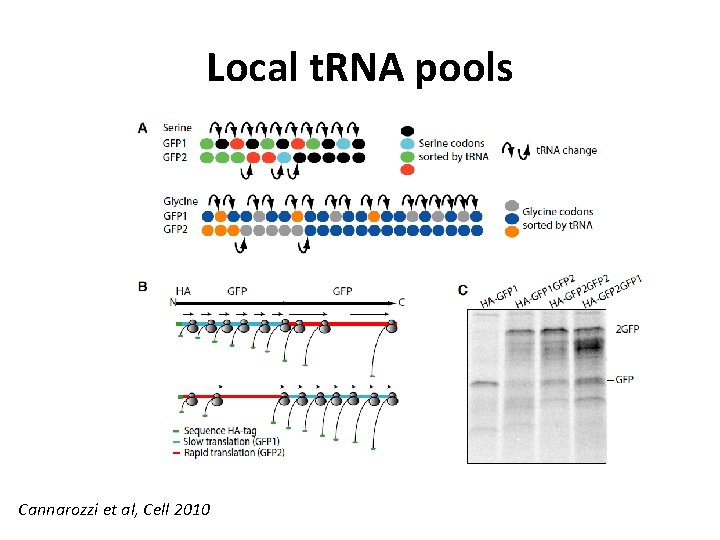
Local t. RNA pools Cannarozzi et al, Cell 2010

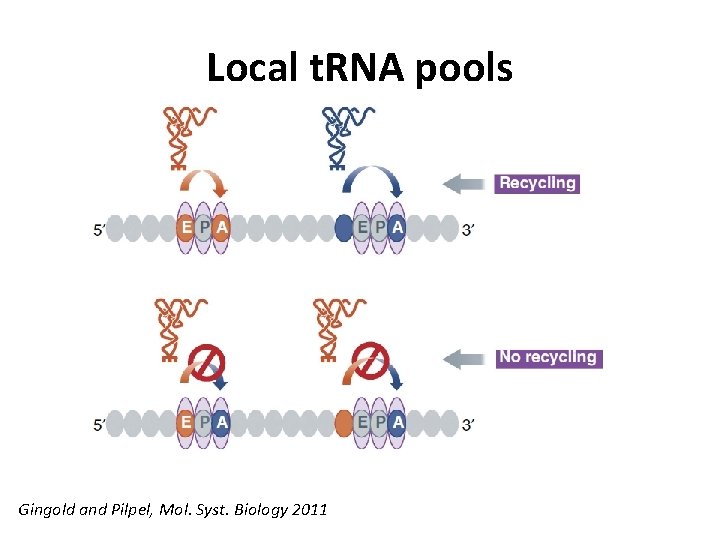
Local t. RNA pools Gingold and Pilpel, Mol. Syst. Biology 2011

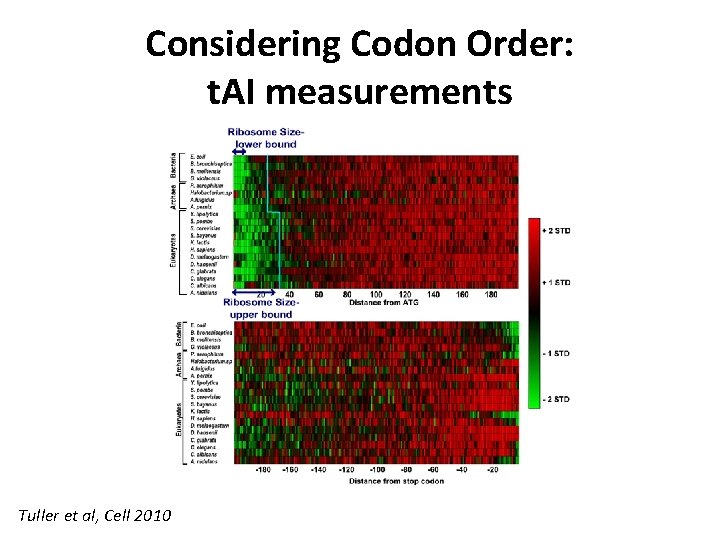
Considering Codon Order: t. AI measurements Tuller et al, Cell 2010

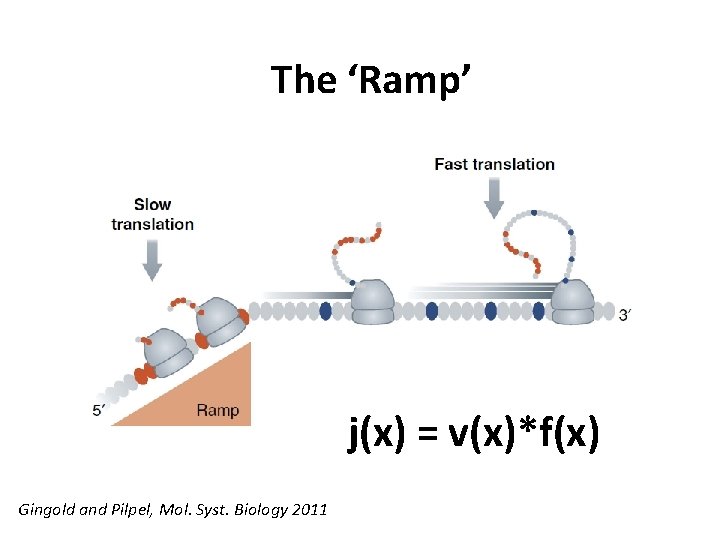
The ‘Ramp’ j(x) = v(x)*f(x) Gingold and Pilpel, Mol. Syst. Biology 2011

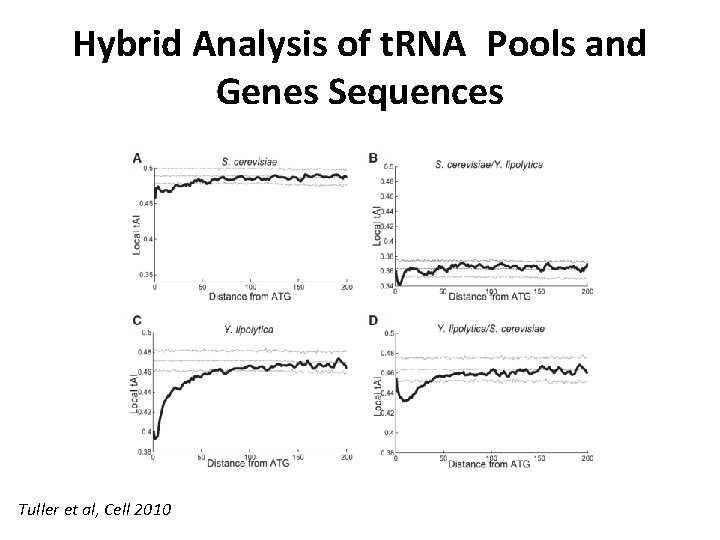
Hybrid Analysis of t. RNA Pools and Genes Sequences Tuller et al, Cell 2010

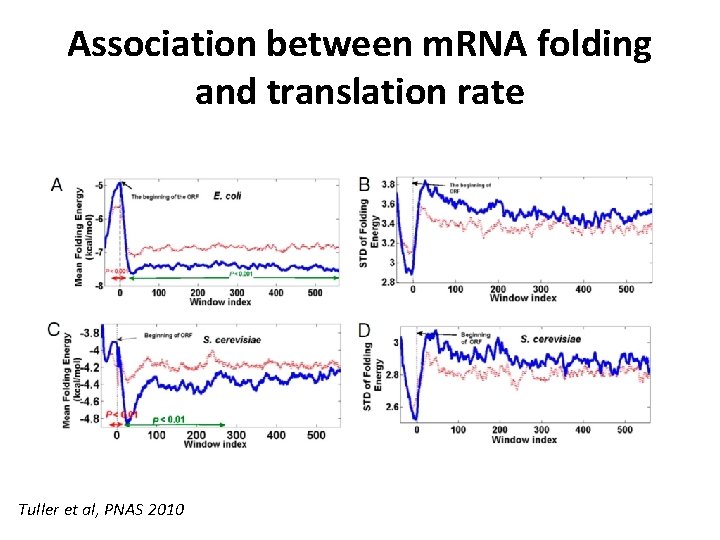
Association between m. RNA folding and translation rate Tuller et al, PNAS 2010

Summary • Most of the genes tend to have lower rate of translation at the beginning of the coding sequence: – Higher ribosome density – Lower t. AI values – Higher m. RNA folding energy • Some genes are transnationally controlled and do not require changes in transcription

More things to discuss • • The effect of translation rate on folding Fidelity Viruses and hosts Mechanistic methods

Thanks