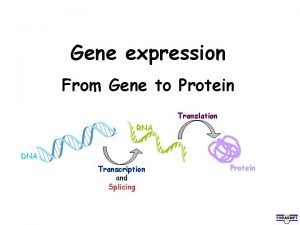
Translation Dr Kevin Ahern Central Dogma m RNA











- Slides: 39

Translation Dr. Kevin Ahern

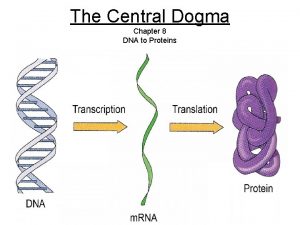
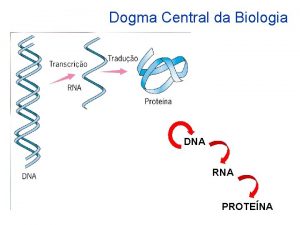
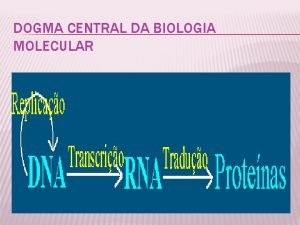
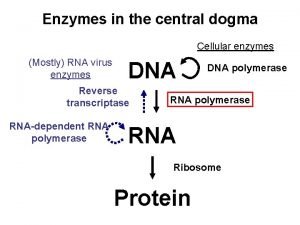
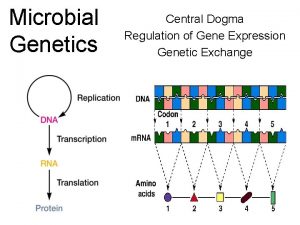
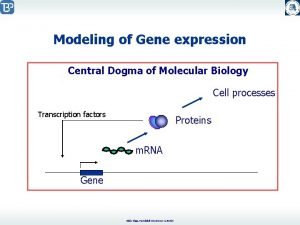
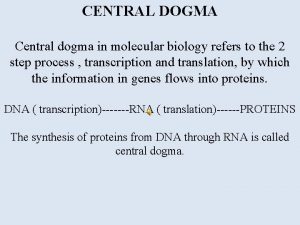
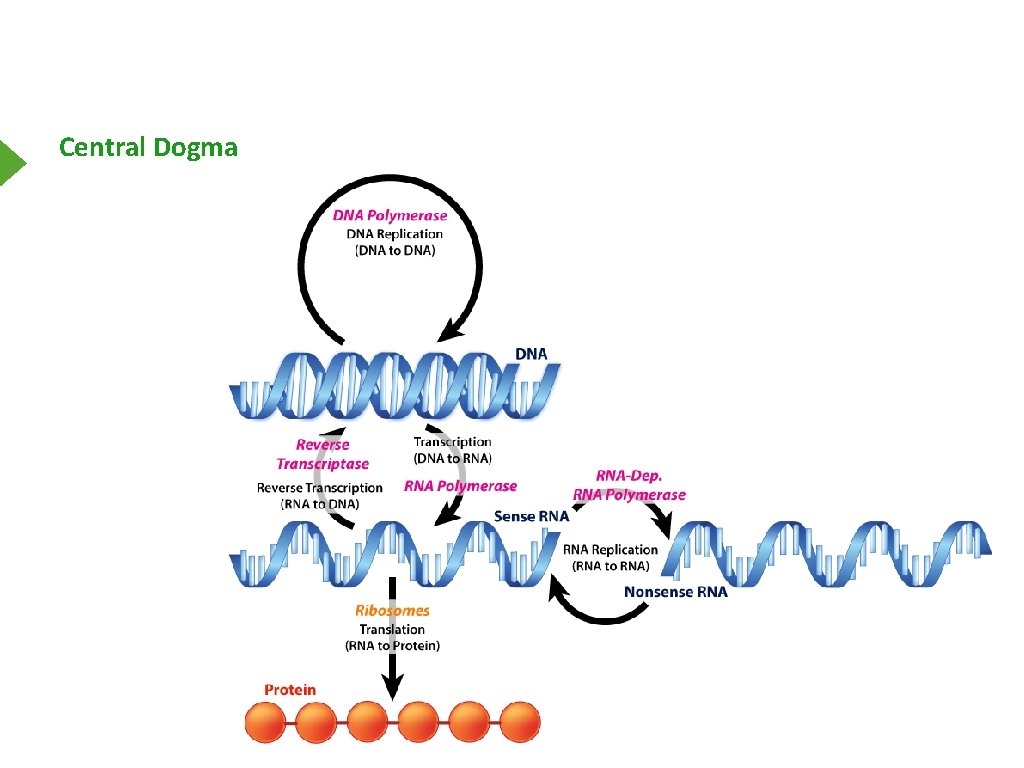
Central Dogma

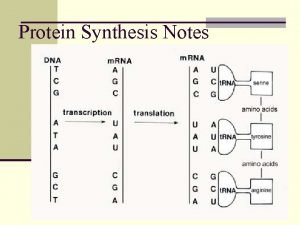
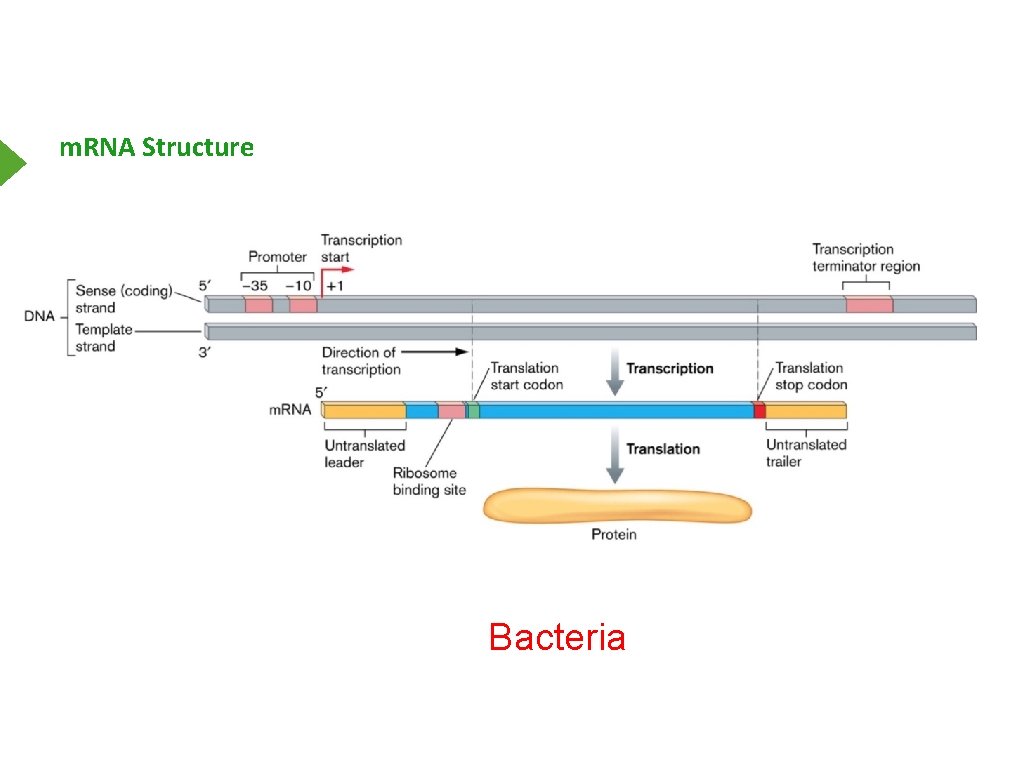
m. RNA Structure Bacteria

Genetic Code Universal Redundant Logical Punctuated 64 Codons 3 Stops 1 Start 2 Unique for AA 59 Redundant

Translation Steps

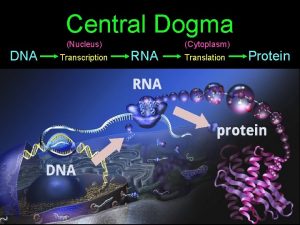
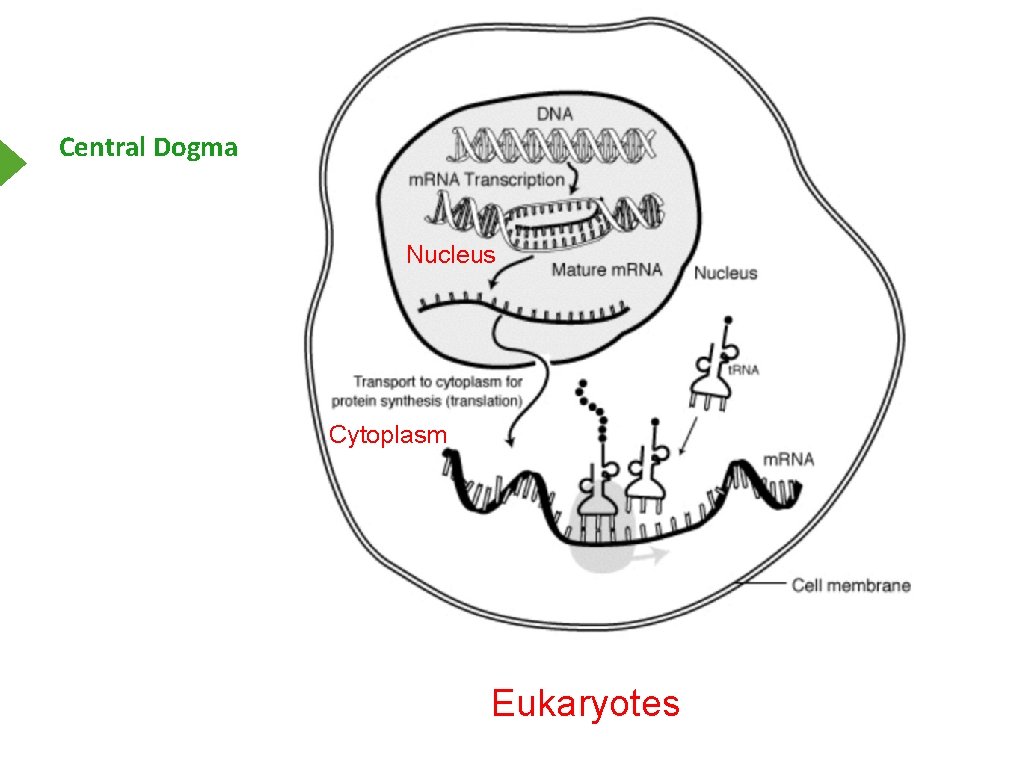
Central Dogma Nucleus Cytoplasm Eukaryotes

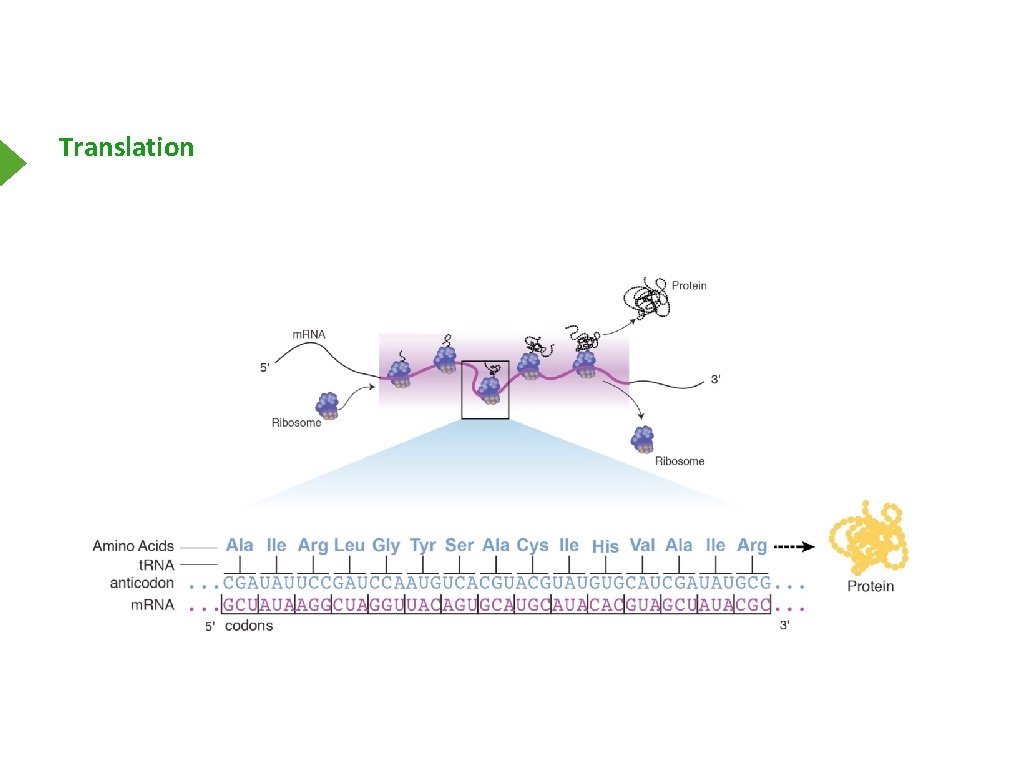
Translation

Simple View of Translation Start Codon Stop Codon

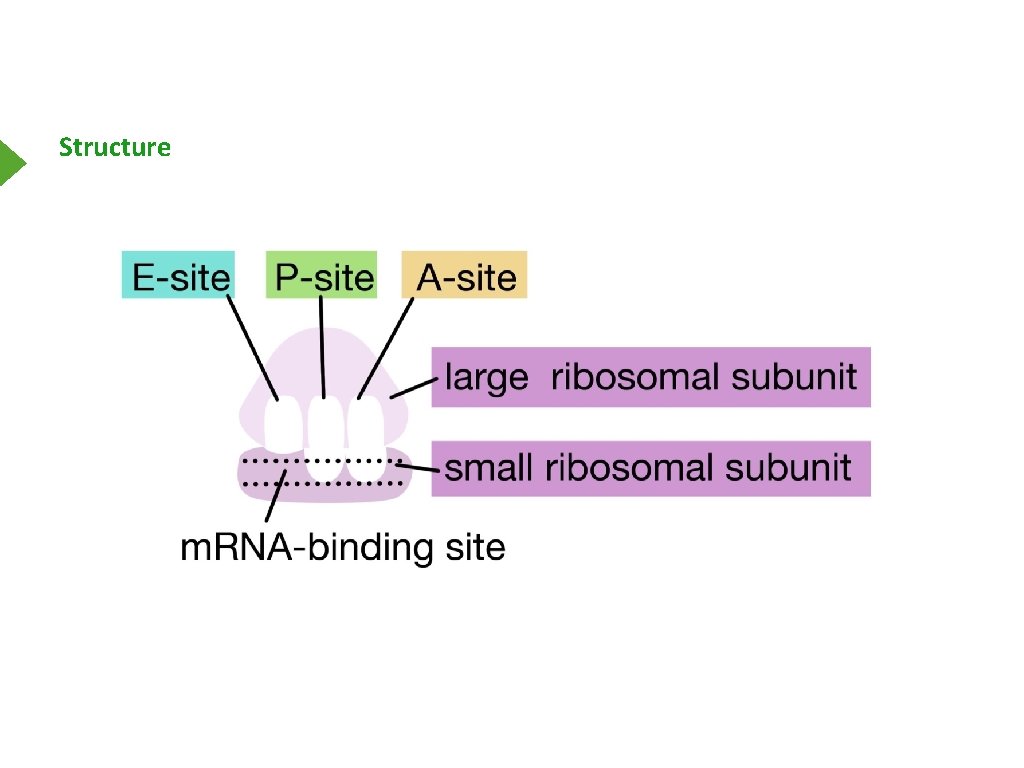
Structure

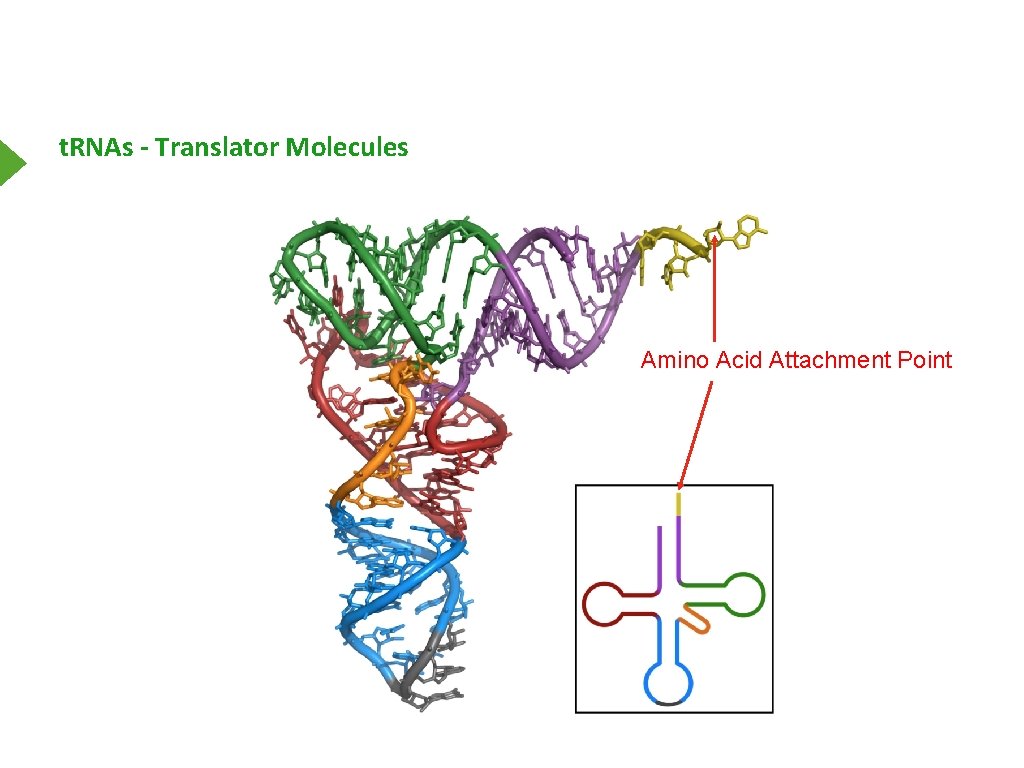
t. RNAs - Translator Molecules Amino Acid Attachment Point

t. RNAs - Translator Molecules

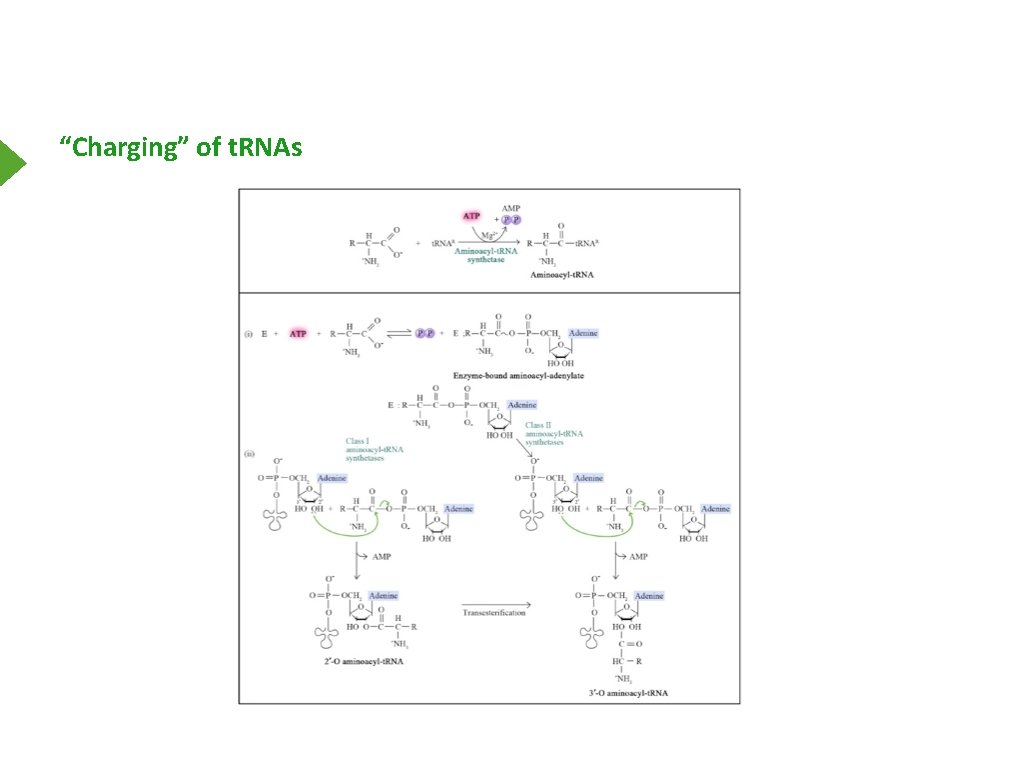
Amino Acid Activation - Charging of t. RNAs Aminoacyl t. RNA-synthetase Type 1 - AA on 2’ OH Type 2 - AA on 3’ OH

“Charging” of t. RNAs

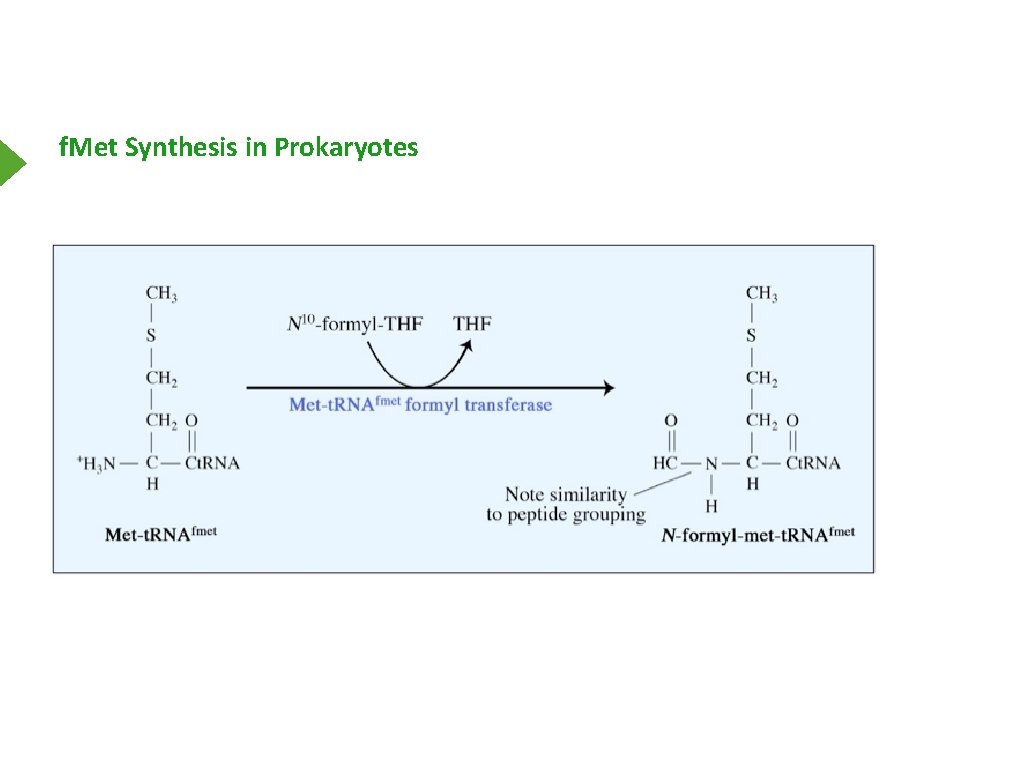
f. Met Synthesis in Prokaryotes

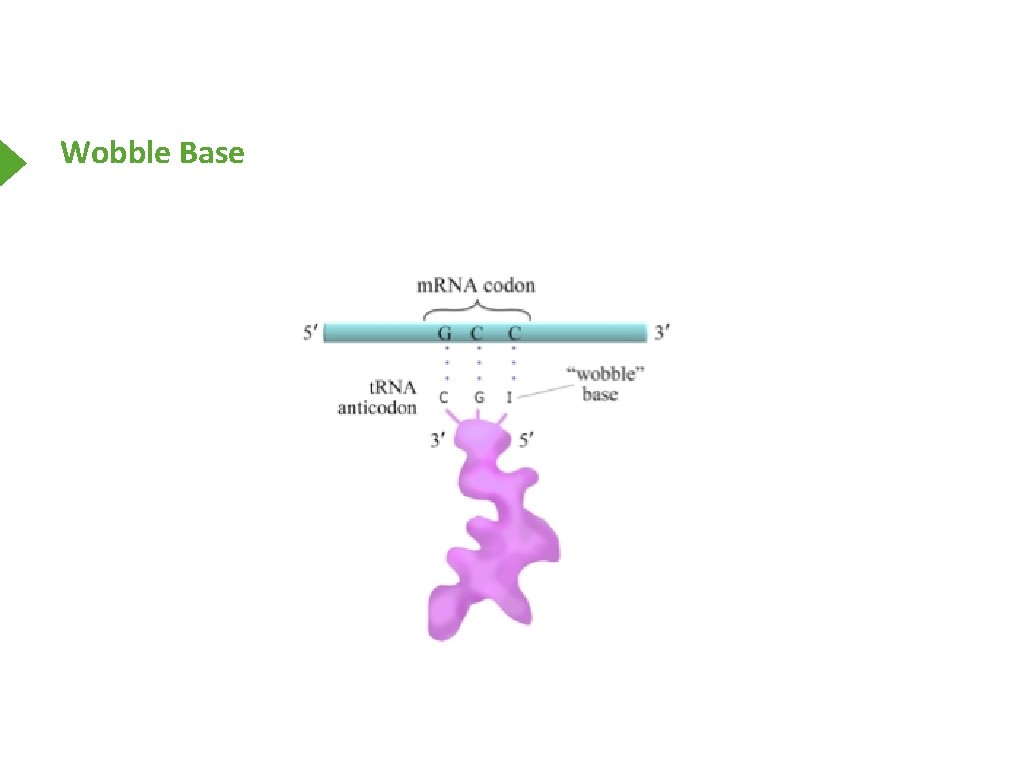
Wobble Base

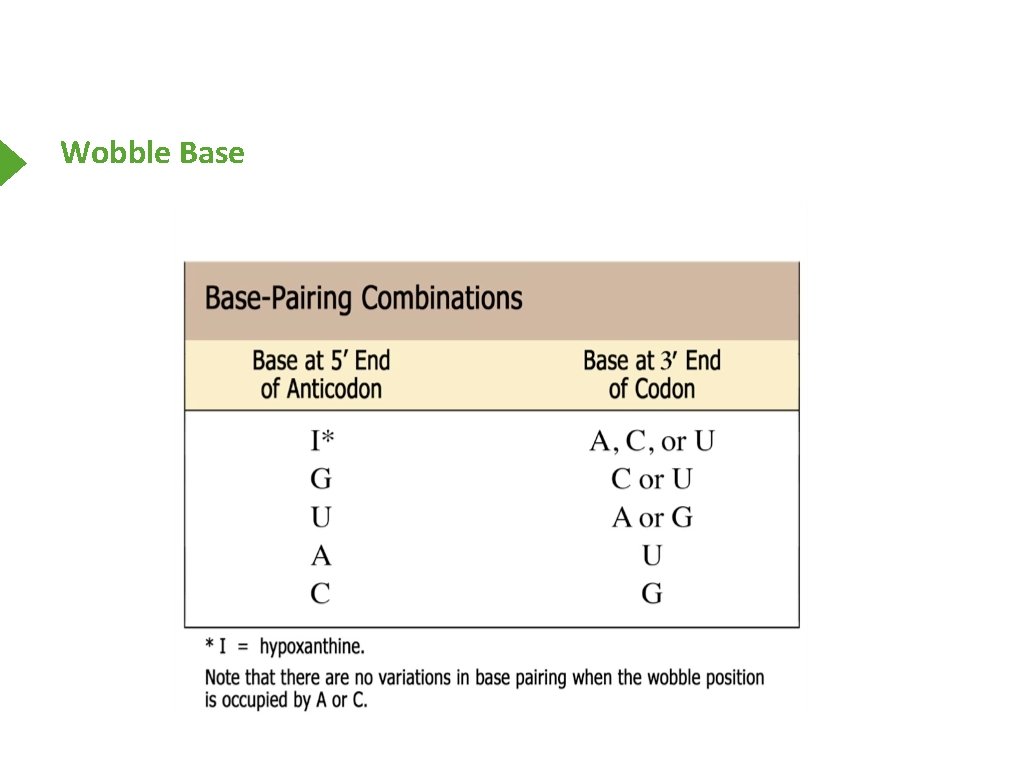
Wobble Base

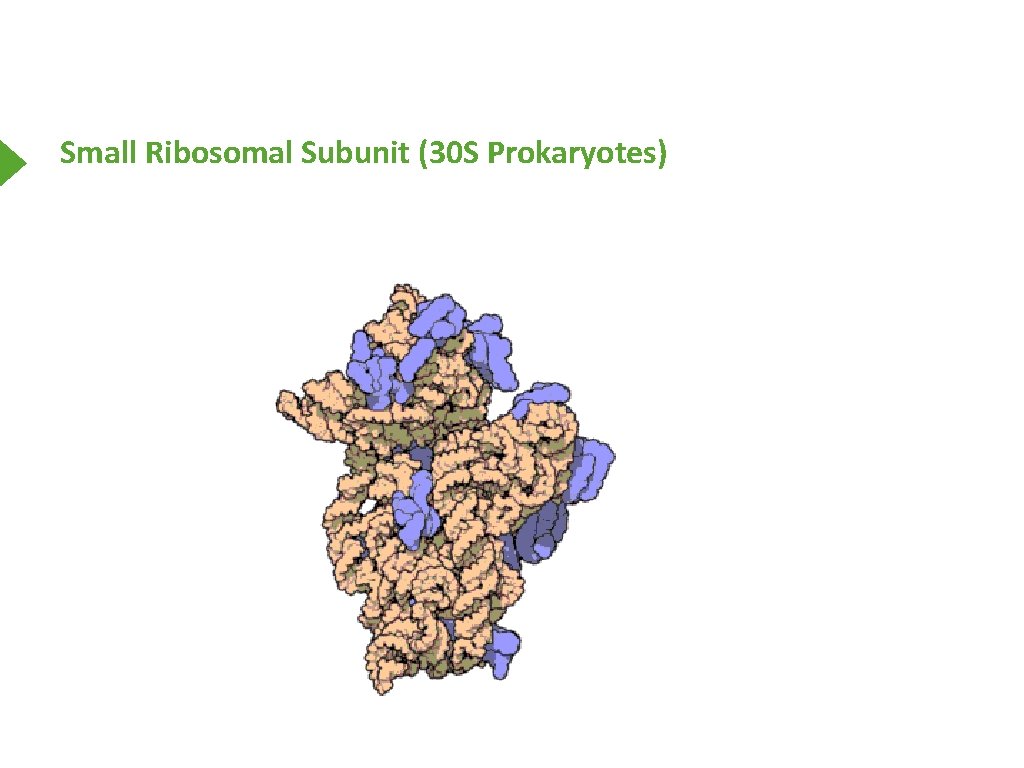
Small Ribosomal Subunit (30 S Prokaryotes)

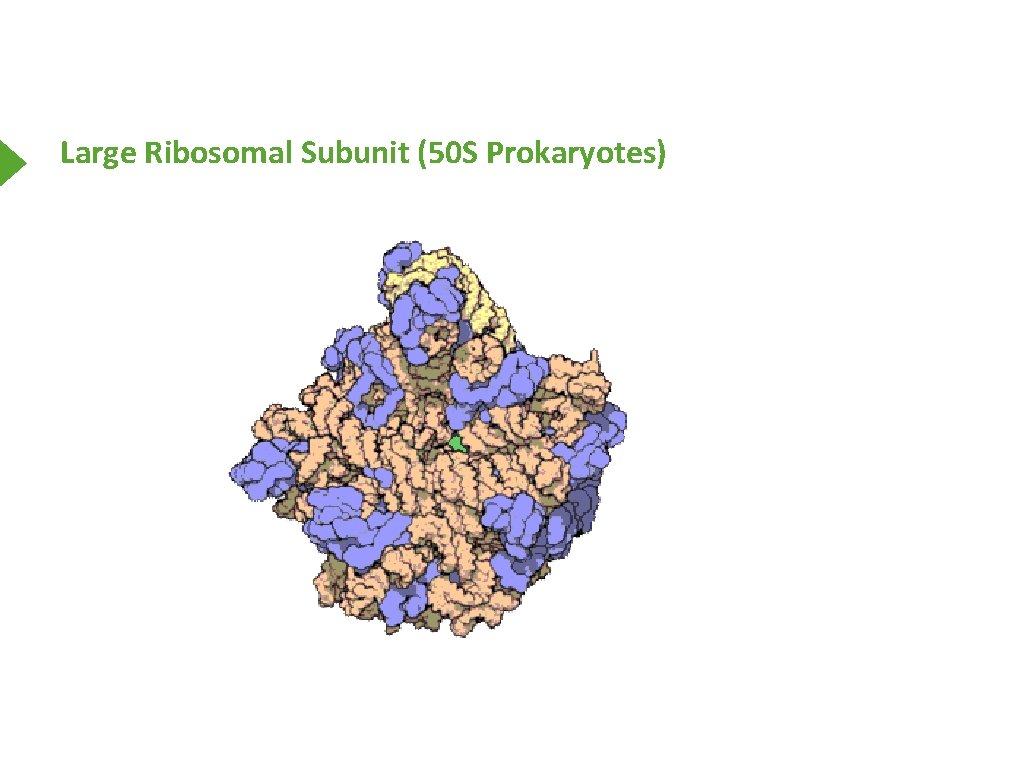
Large Ribosomal Subunit (50 S Prokaryotes)

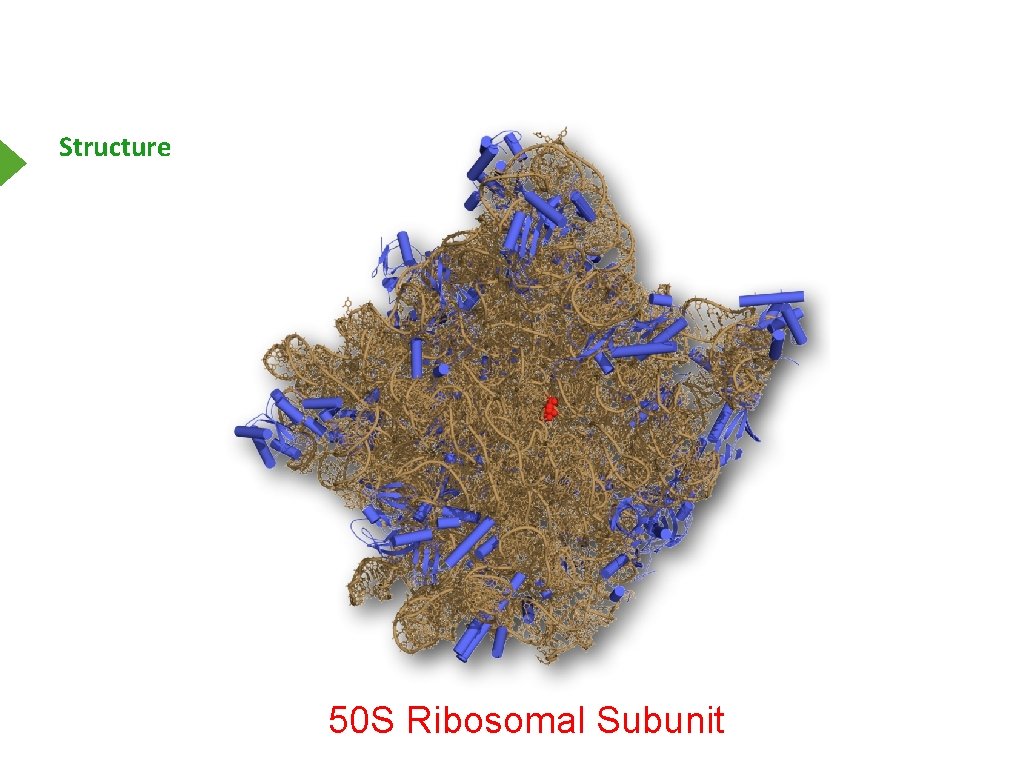
Structure 50 S Ribosomal Subunit

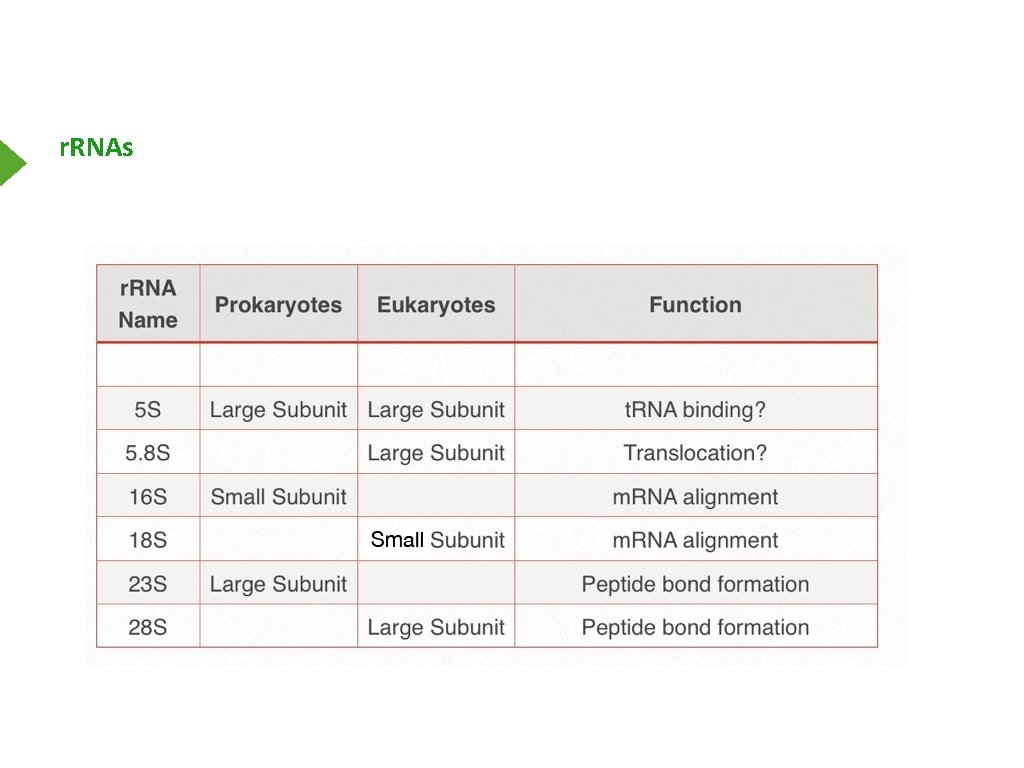
r. RNAs

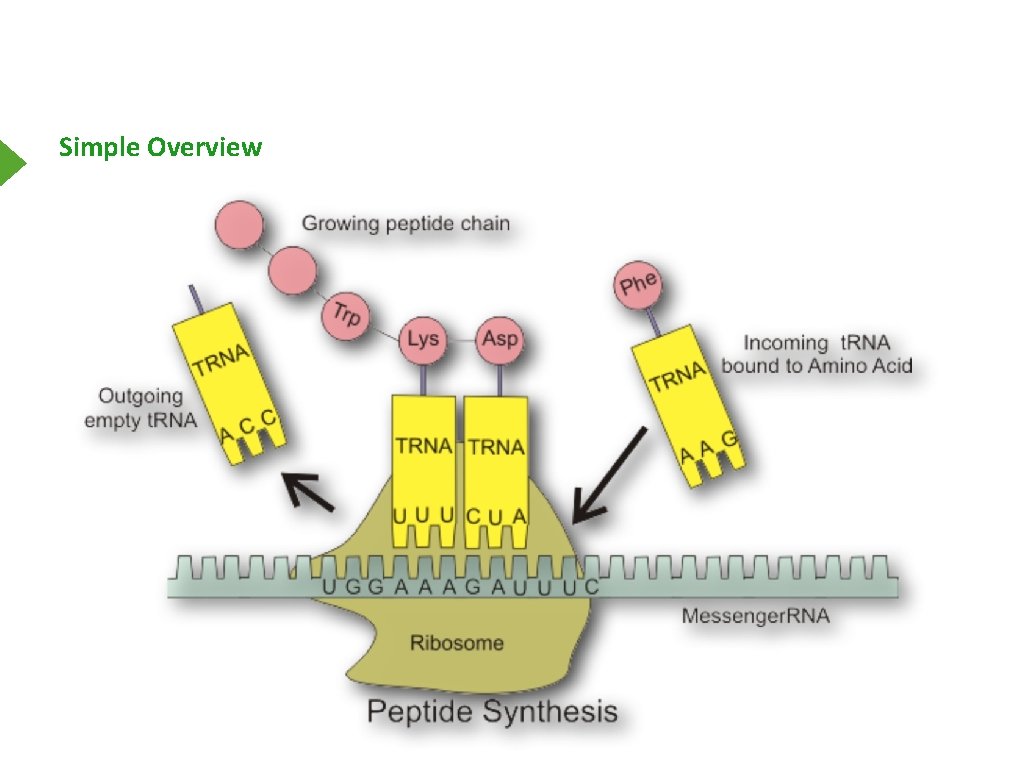
Simple Overview

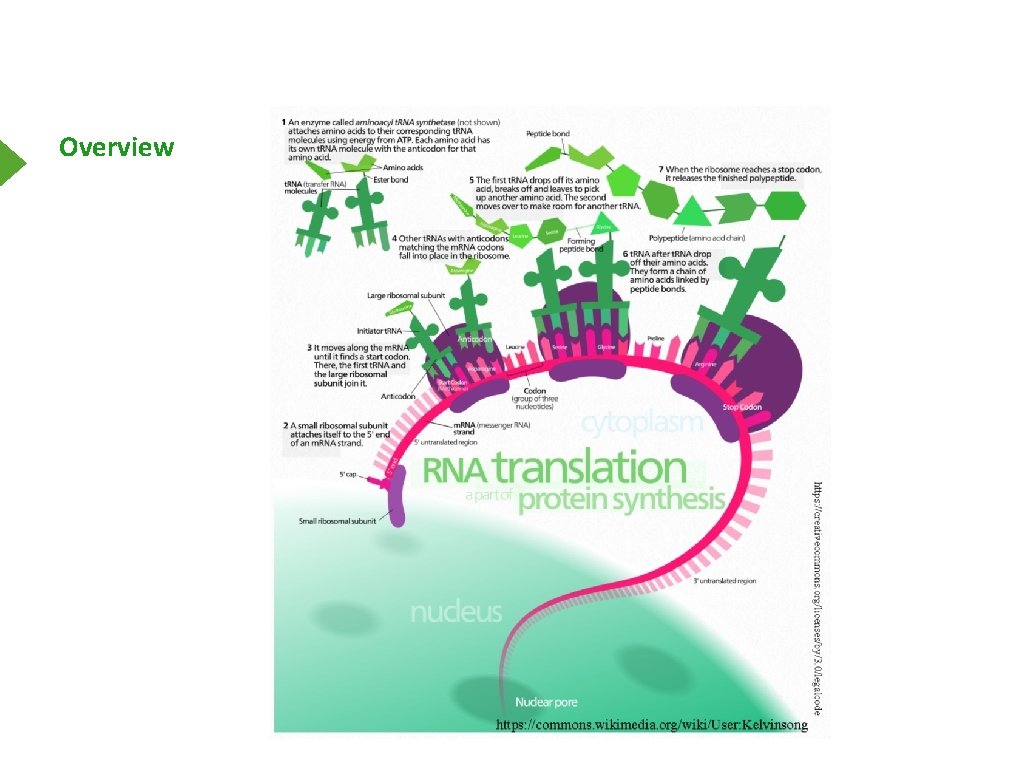
Overview

Prokaryotes - Shine-Dalgarno Sequence

Shine-Dalgarno Sequence 16 S r. RNA Sequence

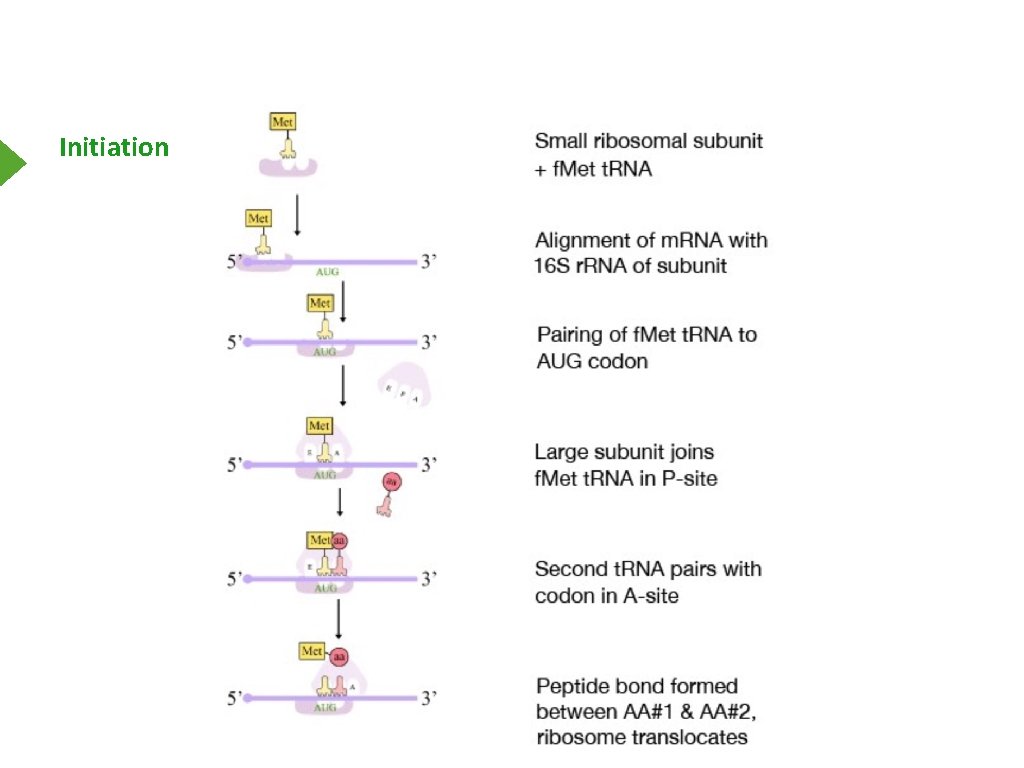
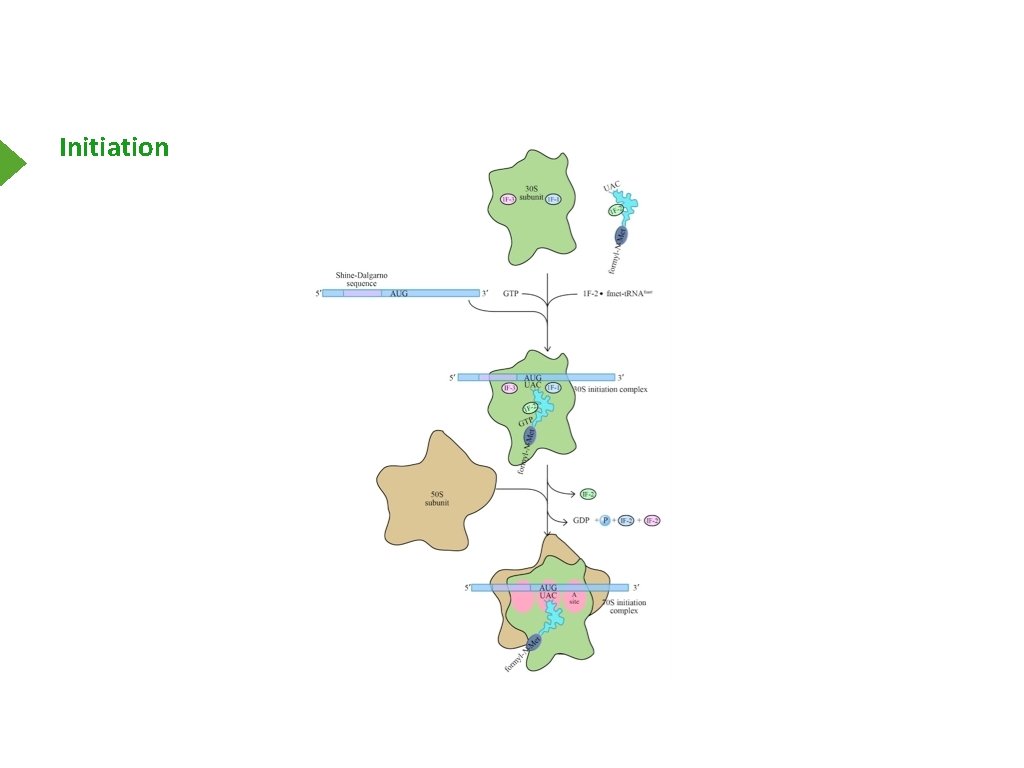
Initiation

Initiation

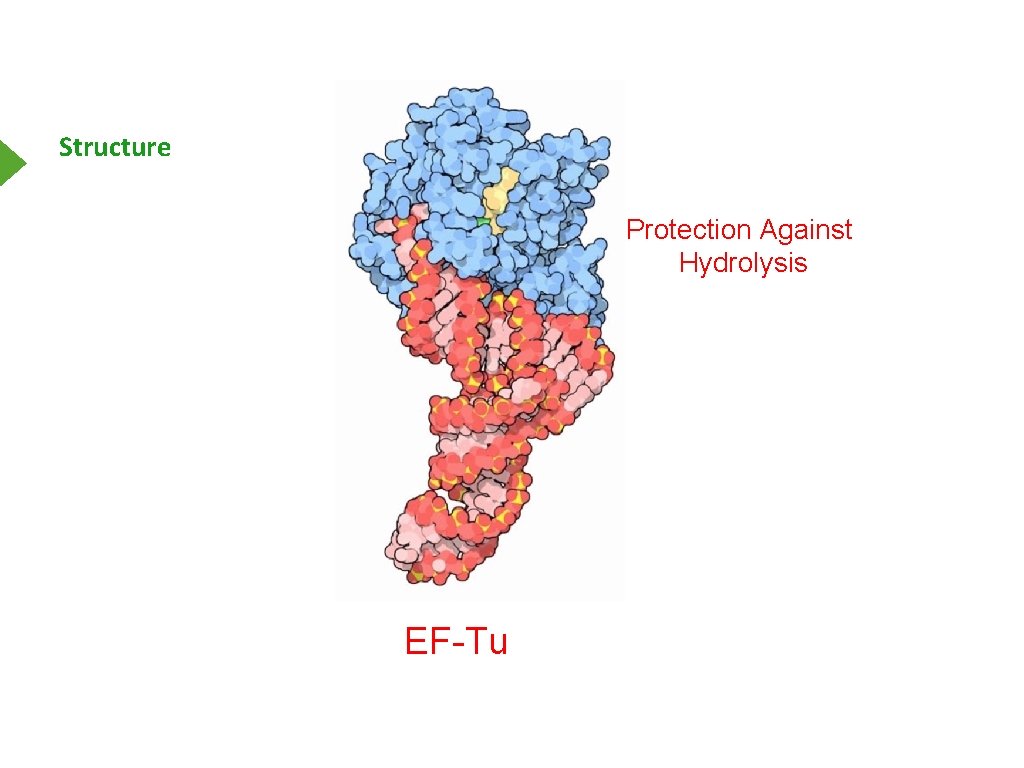
Structure Protection Against Hydrolysis EF-Tu

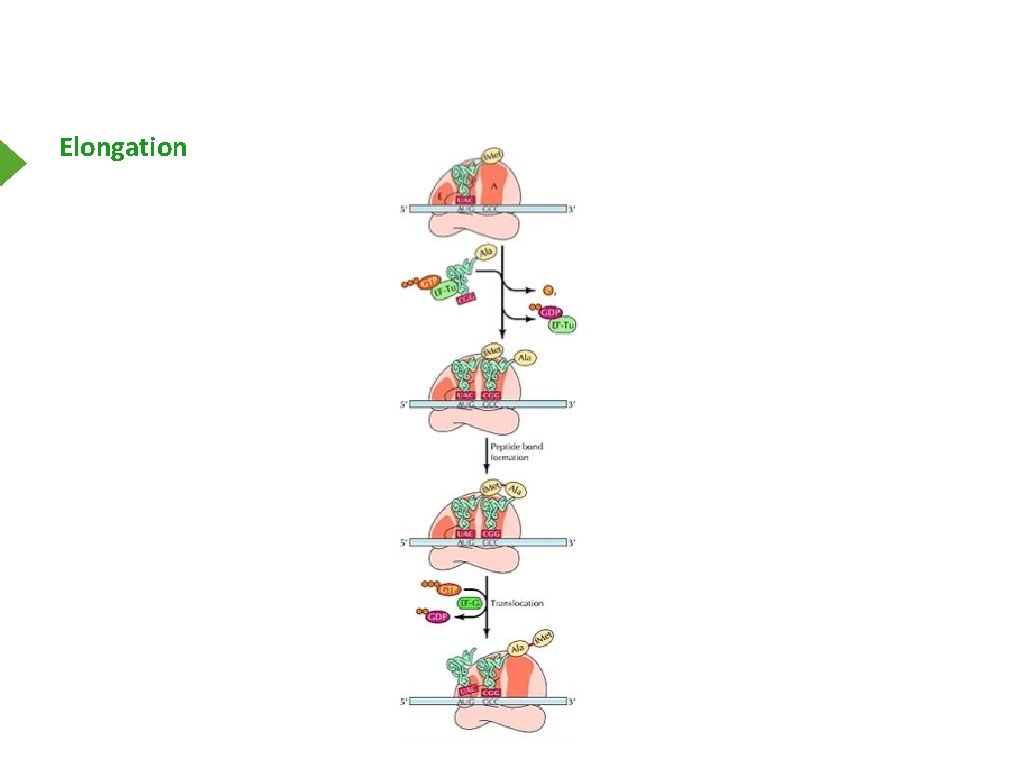
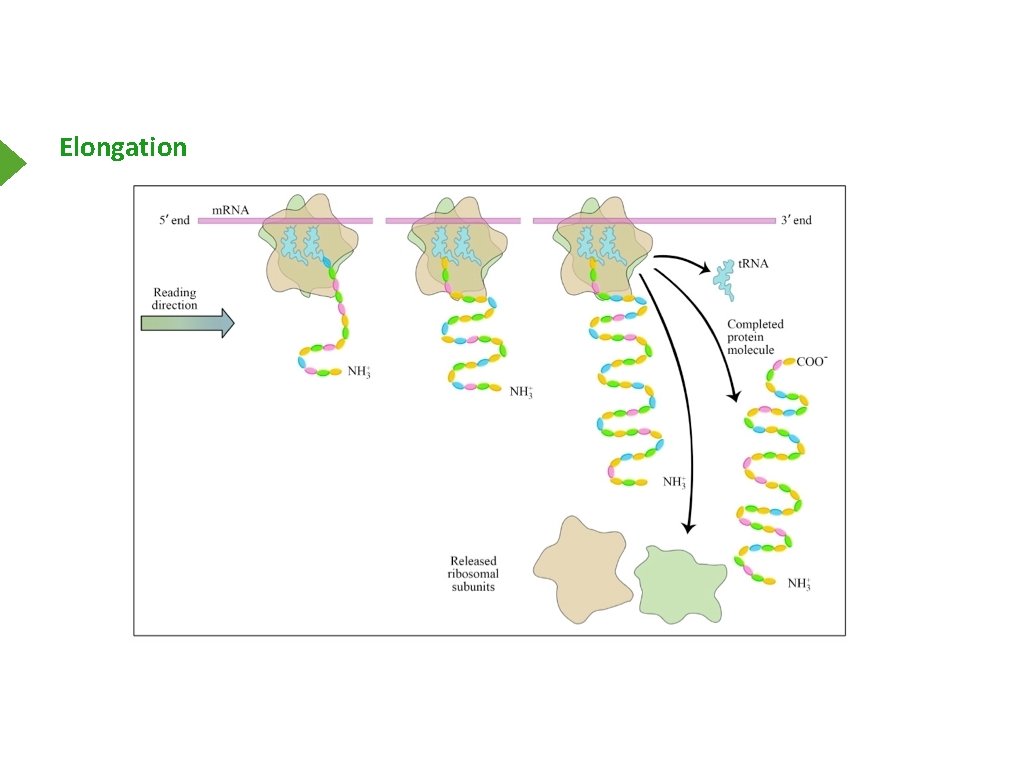
Elongation

Elongation

Termination

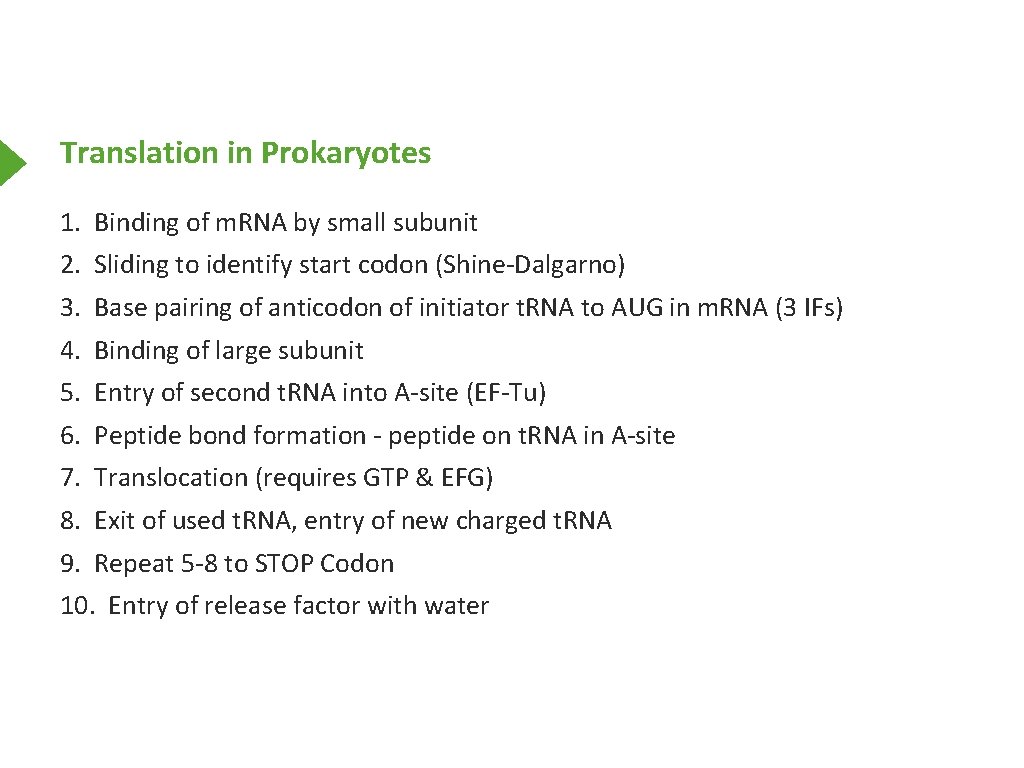
Translation in Prokaryotes 1. Binding of m. RNA by small subunit 2. Sliding to identify start codon (Shine-Dalgarno) 3. Base pairing of anticodon of initiator t. RNA to AUG in m. RNA (3 IFs) 4. Binding of large subunit 5. Entry of second t. RNA into A-site (EF-Tu) 6. Peptide bond formation - peptide on t. RNA in A-site 7. Translocation (requires GTP & EFG) 8. Exit of used t. RNA, entry of new charged t. RNA 9. Repeat 5 -8 to STOP Codon 10. Entry of release factor with water

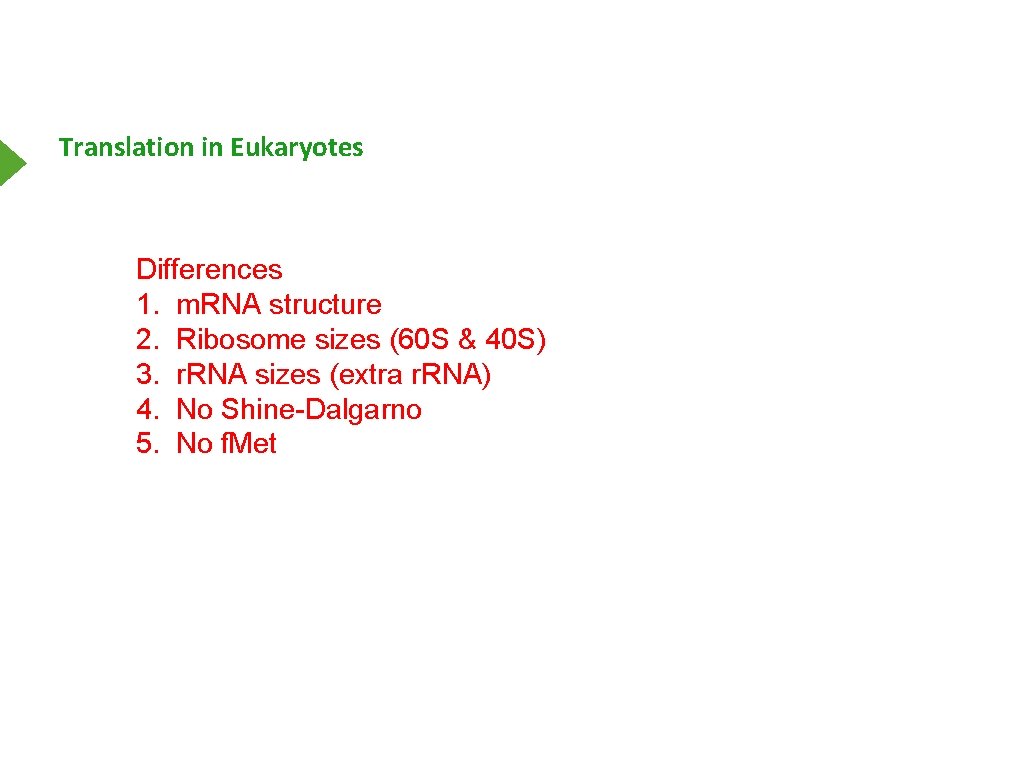
Translation in Eukaryotes Differences 1. m. RNA structure 2. Ribosome sizes (60 S & 40 S) 3. r. RNA sizes (extra r. RNA) 4. No Shine-Dalgarno 5. No f. Met

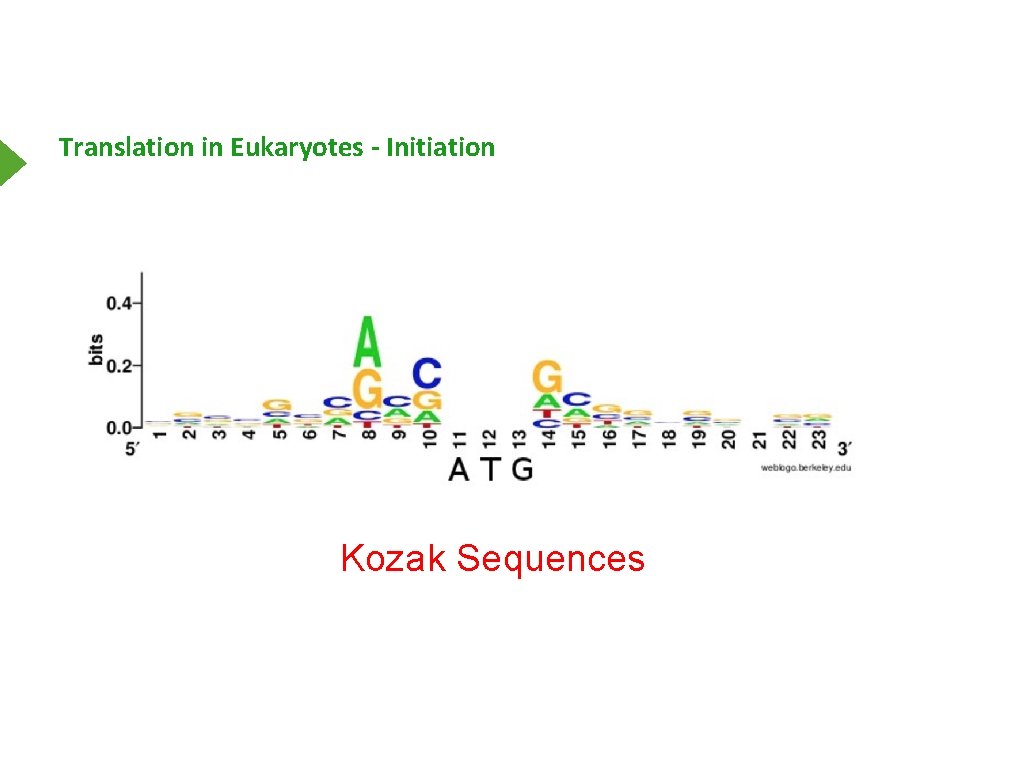
Translation in Eukaryotes - Initiation Kozak Sequences

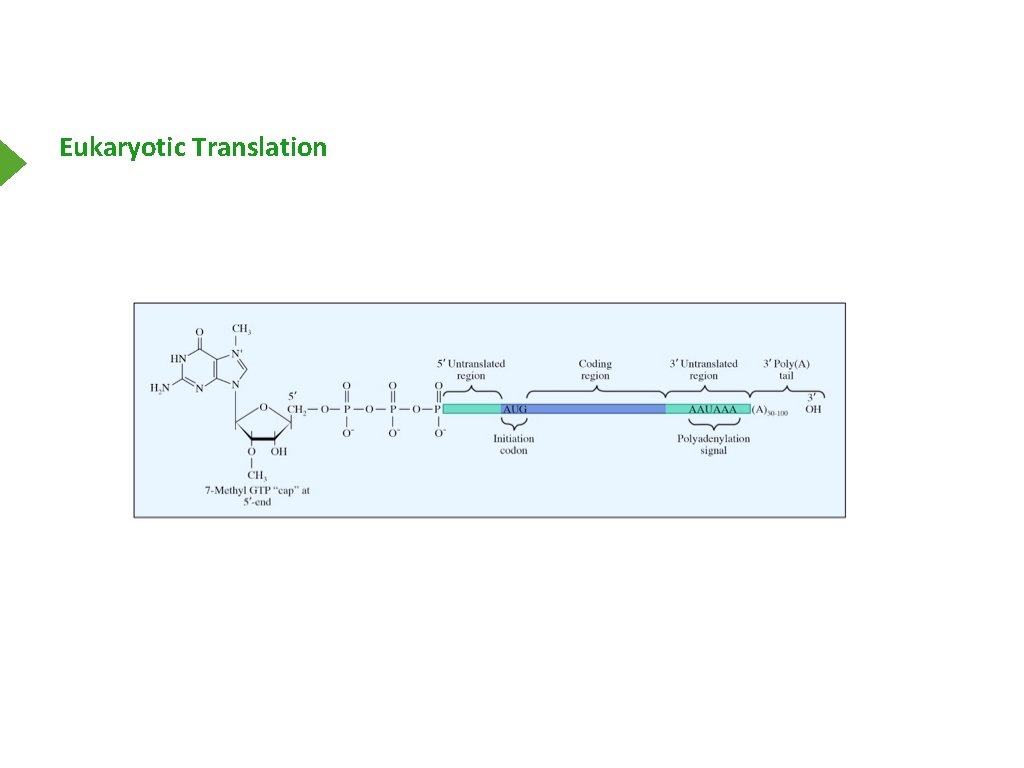
Eukaryotic Translation

Elongation

Eukaryotic Translation Bonding to m. RNA Premature Stopping of Translation Degradation of m. RNA

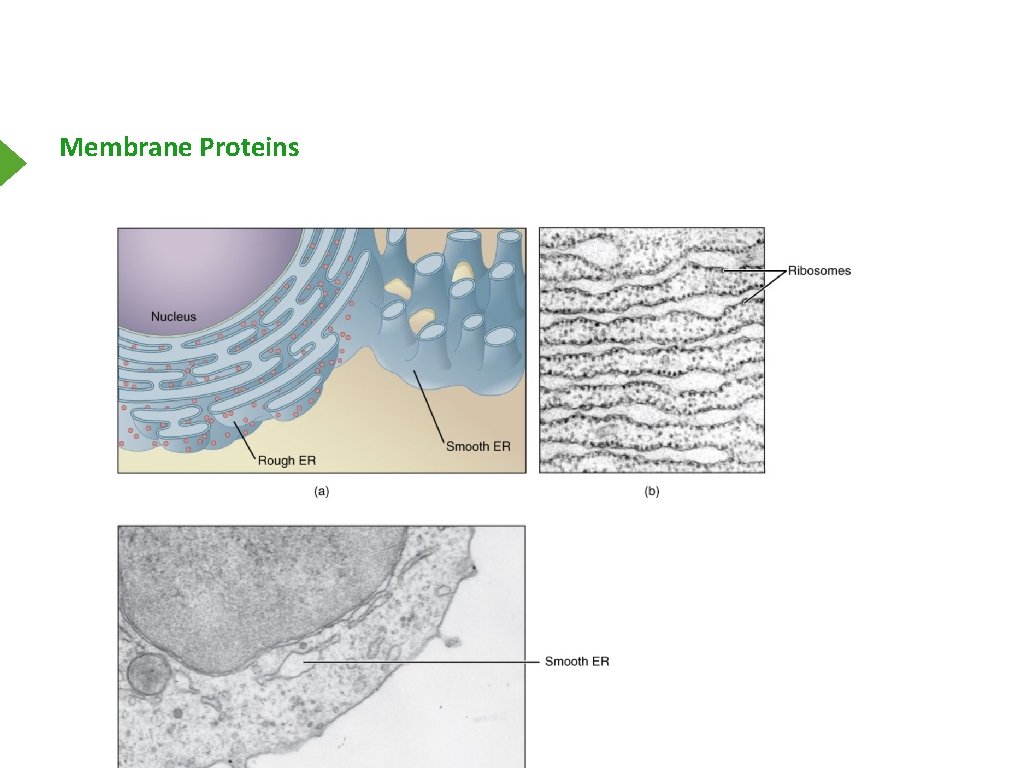
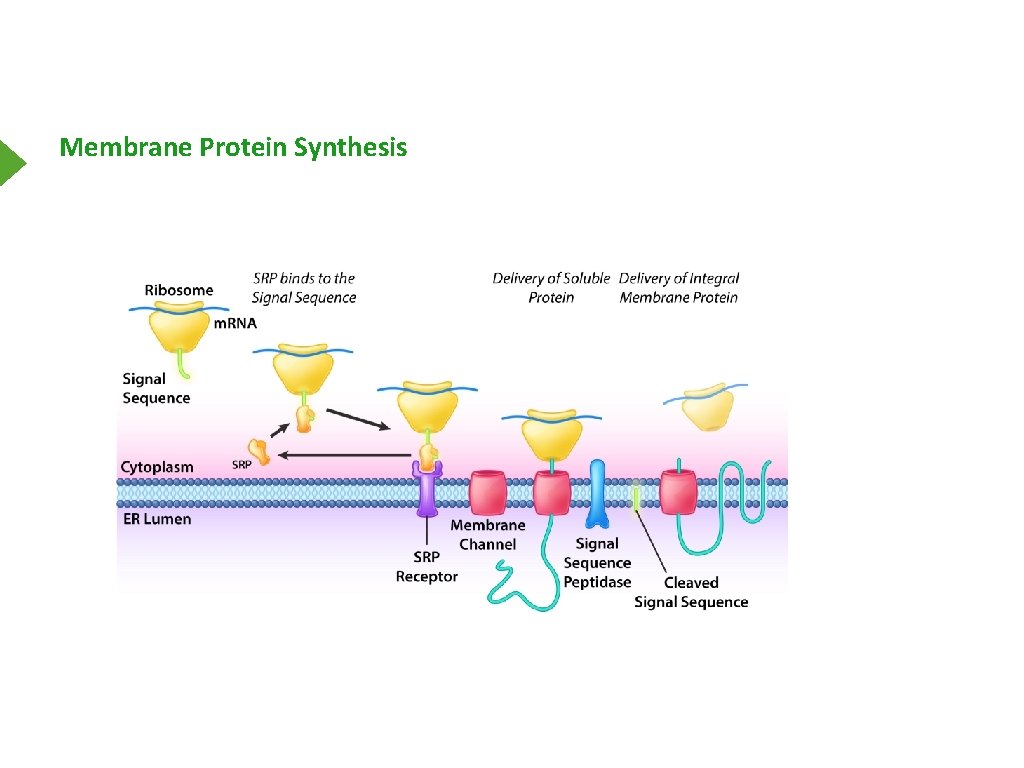
Membrane Proteins

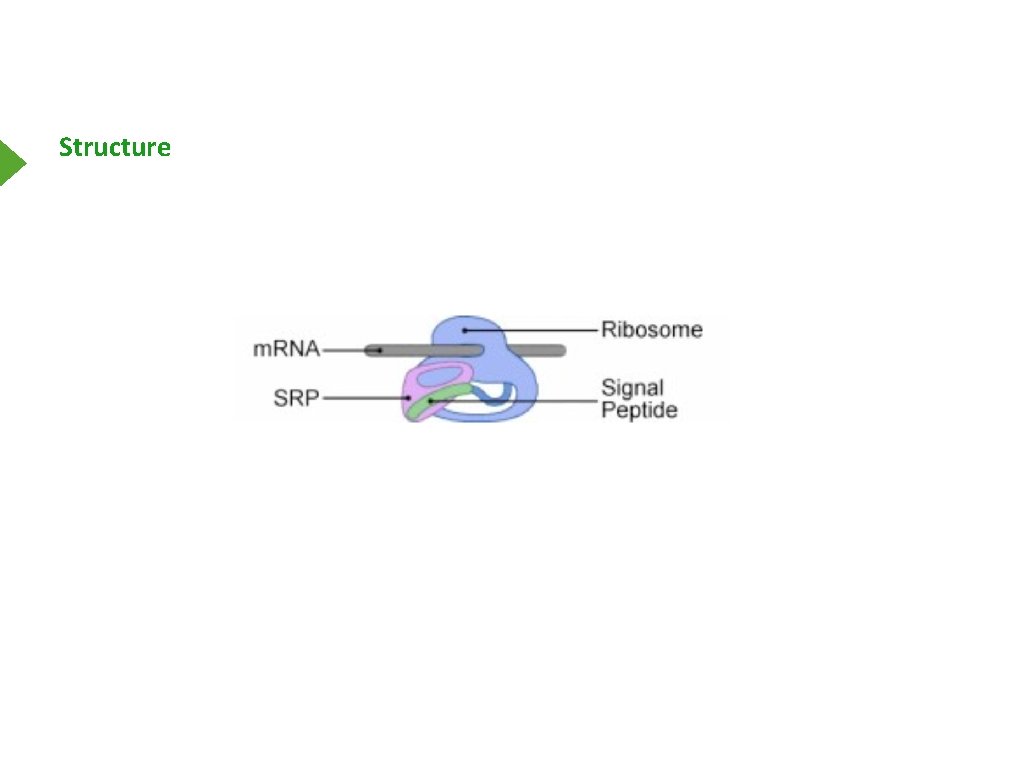
Structure

Membrane Protein Synthesis
 Mutations quiz
Mutations quiz Dna to protein steps
Dna to protein steps Kevin ahern
Kevin ahern Prokaryotic
Prokaryotic Ahern tractors
Ahern tractors Central dogma
Central dogma Central dogma of biology
Central dogma of biology Dogma central
Dogma central Elongation
Elongation Control de la transcripción
Control de la transcripción Central dogma
Central dogma Tradução biologia
Tradução biologia Dogma central
Dogma central Dna central dogma
Dna central dogma Dogma
Dogma Central dogma cartoon
Central dogma cartoon Central dogma diagram
Central dogma diagram Antidepressant side effects
Antidepressant side effects Central dogma
Central dogma Whats the central dogma of biology
Whats the central dogma of biology Semiconservative replication
Semiconservative replication Central dogma
Central dogma Dogma central da biologia
Dogma central da biologia Polirribosomas
Polirribosomas Rna translation
Rna translation Function transformations
Function transformations 10 noun phrases
10 noun phrases Comunicative translation
Comunicative translation Cisco voice translation rule
Cisco voice translation rule Dogma cristologico
Dogma cristologico Translasyon nedir
Translasyon nedir 4 dogmas of mary
4 dogmas of mary Represör
Represör Rna transfer
Rna transfer Dogma of biology
Dogma of biology Dogma definition
Dogma definition Isaac repentance dogma
Isaac repentance dogma Dogma
Dogma Distributed systems middleware
Distributed systems middleware George boole
George boole