Transgene design Each transgene contains a 1 promoter




- Slides: 16

Transgene design Each transgene contains a: 1. promoter, 2. an intron, 3. a protein coding sequence 4. a reporter gene and 5. transcriptional stop sequence. Ø These elements are typically assembled in a bacterial plasmid. Ø Sequences are usually chosen from previous transgenes with proven function. Ø Prokaryotic sequences removed before injection into the nucleus of a zygote.

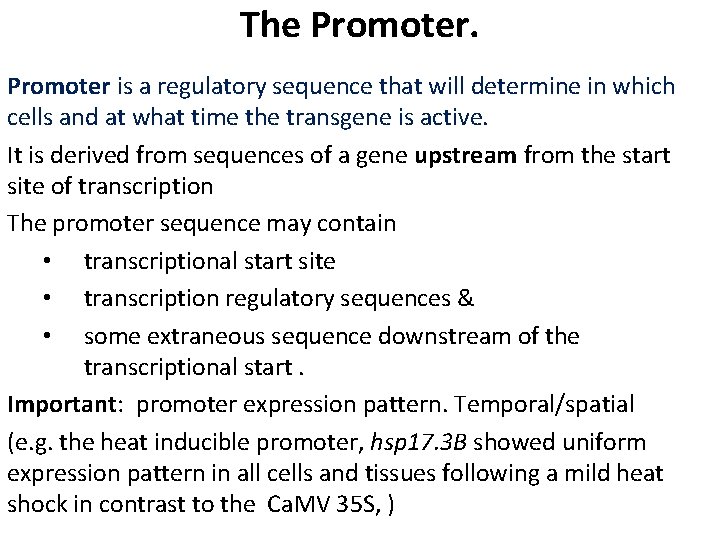
The Promoter is a regulatory sequence that will determine in which cells and at what time the transgene is active. It is derived from sequences of a gene upstream from the start site of transcription The promoter sequence may contain • transcriptional start site • transcription regulatory sequences & • some extraneous sequence downstream of the transcriptional start. Important: promoter expression pattern. Temporal/spatial (e. g. the heat inducible promoter, hsp 17. 3 B showed uniform expression pattern in all cells and tissues following a mild heat shock in contrast to the Ca. MV 35 S, )

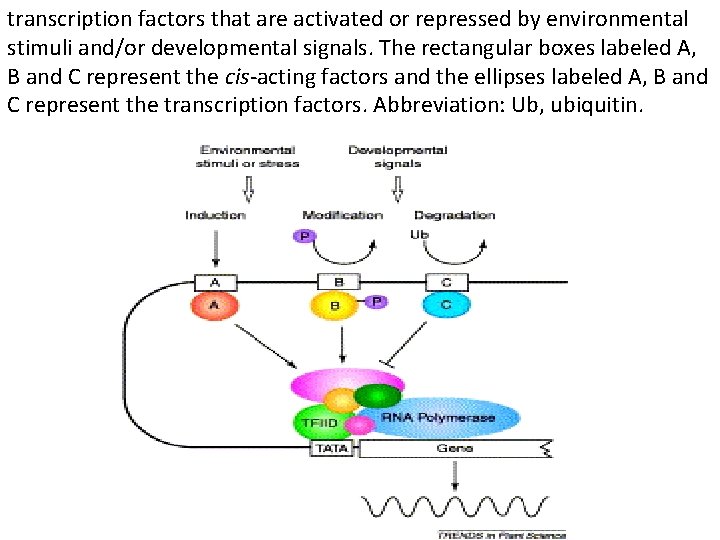
transcription factors that are activated or repressed by environmental stimuli and/or developmental signals. The rectangular boxes labeled A, B and C represent the cis-acting factors and the ellipses labeled A, B and C represent the transcription factors. Abbreviation: Ub, ubiquitin.

The promoter DNA is the progressively digested from it's 5' end & reporter gene activity. The difference in activity between different promoter fragment lengths indicates the presence of a regulatory element in the section of DNA which was deleted.

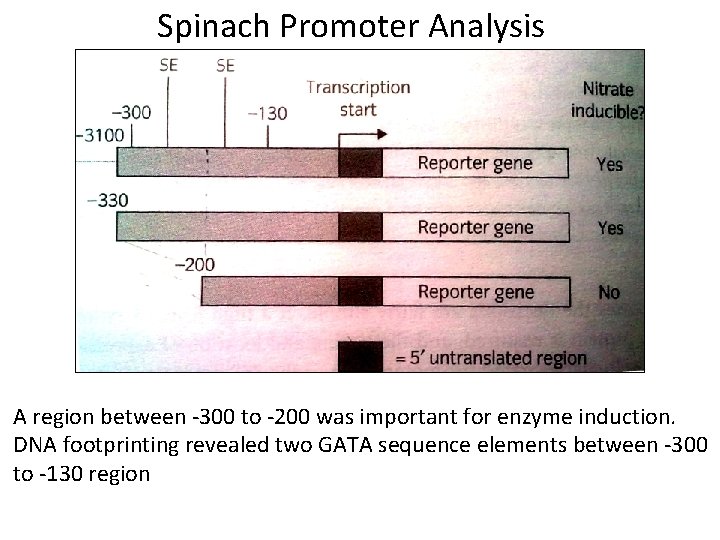
Spinach Promoter Analysis A region between -300 to -200 was important for enzyme induction. DNA footprinting revealed two GATA sequence elements between -300 to -130 region

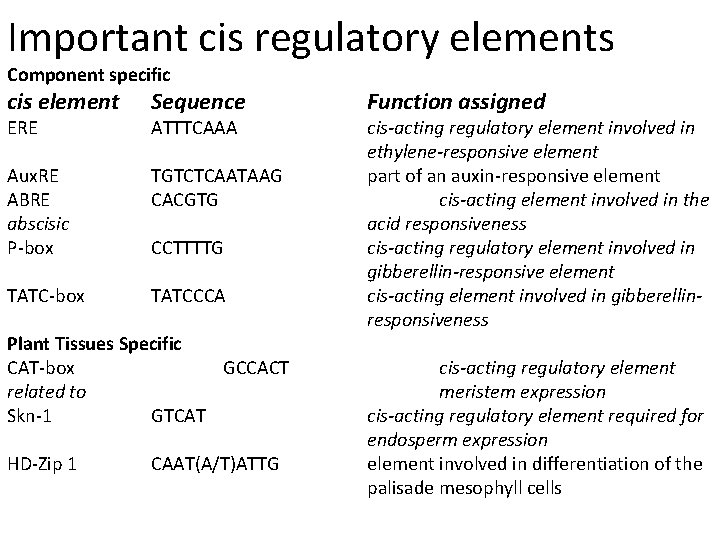
Important cis regulatory elements Component specific cis element Sequence Aux. RE ABRE abscisic P-box TGTCTCAATAAG CACGTG TATC-box TATCCCA ERE ATTTCAAA CCTTTTG Plant Tissues Specific CAT-box GCCACT related to Skn-1 GTCAT HD-Zip 1 CAAT(A/T)ATTG Function assigned cis-acting regulatory element involved in ethylene-responsive element part of an auxin-responsive element cis-acting element involved in the acid responsiveness cis-acting regulatory element involved in gibberellin-responsive element cis-acting element involved in gibberellinresponsiveness cis-acting regulatory element meristem expression cis-acting regulatory element required for endosperm expression element involved in differentiation of the palisade mesophyll cells

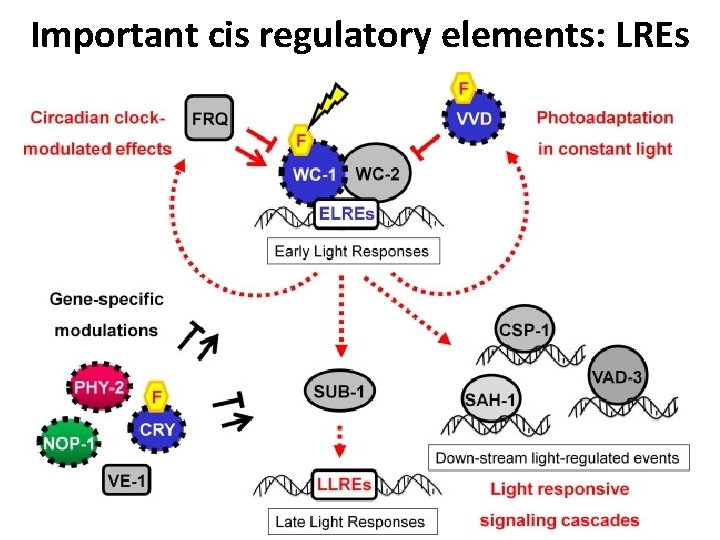
Important cis regulatory elements: LREs

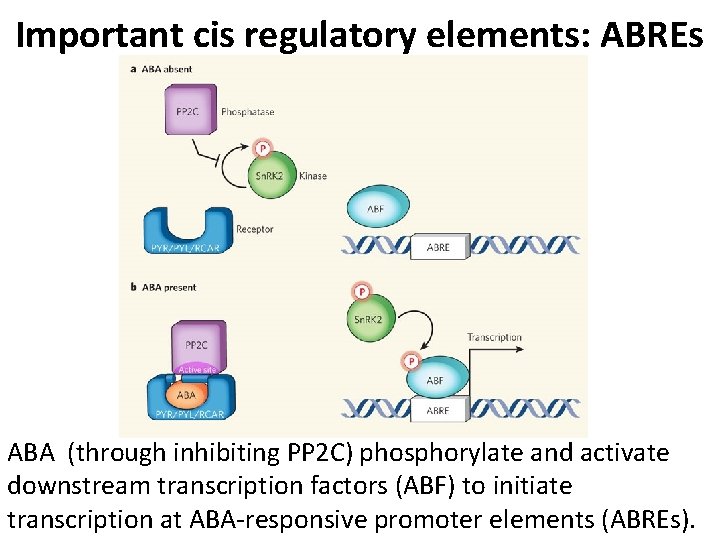
Important cis regulatory elements: ABREs ABA (through inhibiting PP 2 C) phosphorylate and activate downstream transcription factors (ABF) to initiate transcription at ABA-responsive promoter elements (ABREs).

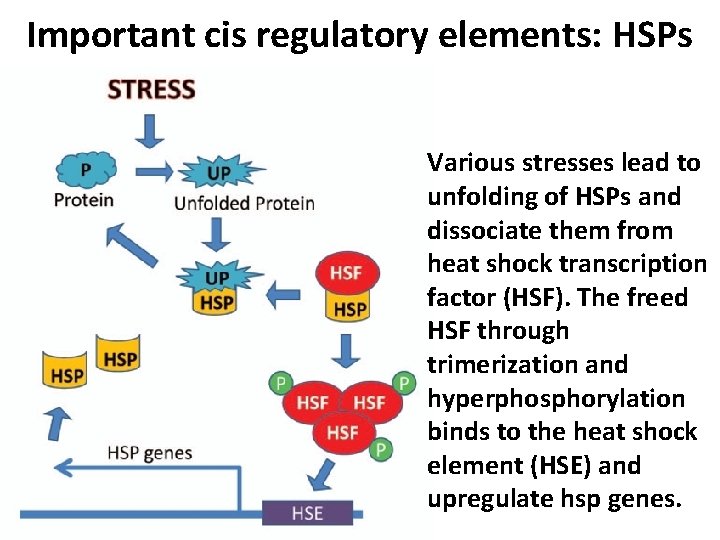
Important cis regulatory elements: HSPs Various stresses lead to unfolding of HSPs and dissociate them from heat shock transcription factor (HSF). The freed HSF through trimerization and hyperphosphorylation binds to the heat shock element (HSE) and upregulate hsp genes.

Synthetic promoters Promoters are needed to work : • • • under a desired condition, at a desired strength and in a specified tissue Two ways to the synthesize promoters: • One through bioinformatical model and database (HEARTBEAT) for promoters responsive to userdefined inputs. • Second through biochemical method to synthesize promoter libraries.

Synthetic promoters core promoter or minimal promoter contains: 1. TATA box, 2. transcription start site or CAP site May or may not have Detectable activity Other elements of promoter regions include: 1. INR (initiator) sequences near the transcription start site; 2. enhancers; and 3. upstream elements (e. g. a range of cis-acting elements are needed to mediate local gene expression in plant (boxes W 1, W 2, GCC, JERE, S, Gst 1, and D after pathogen attack). Synthetic promoters with specialized functions have also been developed.

The reporter (protein coding sequence). Transgenes must contain a valid protein coding sequence (CDS) derived from the c. DNA. The CDS must contain 1. a translational start codon (ATG) 2. translational stop codon & 3. a Kozak sequence (5′ UTR that allows ribosomes to recognize the initiator codon)upstream of the start codon. • • • The ideal Kozak sequence is GCCGCCACC. Extra linker sequences (for RE) (devoid of start codons). 5’ and 3’ non-translated sequences from the protein coding transcript should be avoided as much as possible, since these may contain regulatory elements controlling translation or m. RNA stability.

Introns 1. Integration site: transgene expression may be more active in one site than another (factors ? ). 2. Inclusion of an intron results in a significantly greater percentage of active transgenes. (e. g. 6/7 transgenes with an intron had detectable activity, while 2/5 identical constructs without an intron had detectable weaker expression)

transcriptional stop signal Each transgene must also contain a transcriptional stop signal to match the start signal typically included in the promoter. Eukaryotic transcriptional stop signals include • a poly. A addition sequence (AAAUAA) & • hundreds of downstream nucleotides whose function is not clearly understood. Commonly used introns are -rabbit β-globin or -SV 40 (Simian vacuolating virus 40 ) & stop sequences are from -SV 40 or -human growth hormone.

Editing the transgene Transgene sould be 1. linear 2. Free from bacterial ori and 3. Free from prokaryotic sequences.

Linker and extra sequences Typical cloning methods will have • Restriction endonuclease sites or • Plasmid polylinker sequences These sequence must be free of • translational start or stop sites & • unwanted functional elements like enhancers & promoters.