Transformations in Statistical Analysis Outline Assumptions of linear
- Slides: 21

Transformations in Statistical Analysis Outline • Assumptions of linear statistical models. • Types of Transformations • Alternatives to Transformations Model Assumptions • • Effect addivitity Normality Homoscedasticity Independence 1

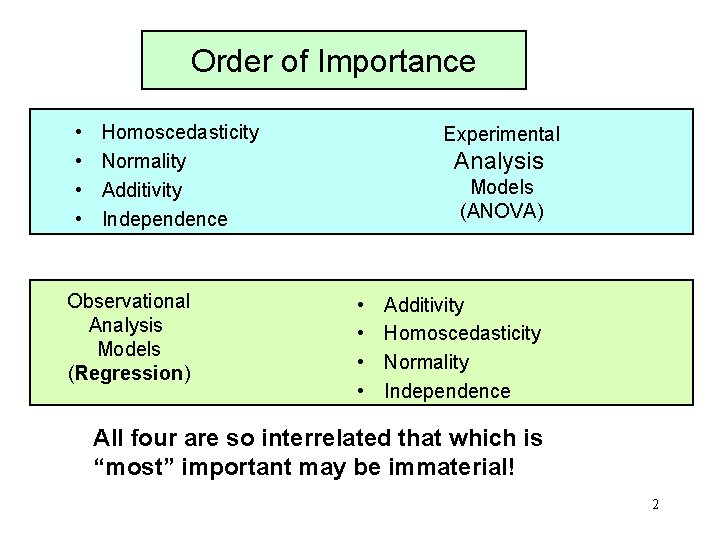
Order of Importance • • Homoscedasticity Normality Additivity Independence Observational Analysis Models (Regression) Experimental Analysis Models (ANOVA) • • Additivity Homoscedasticity Normality Independence All four are so interrelated that which is “most” important may be immaterial! 2

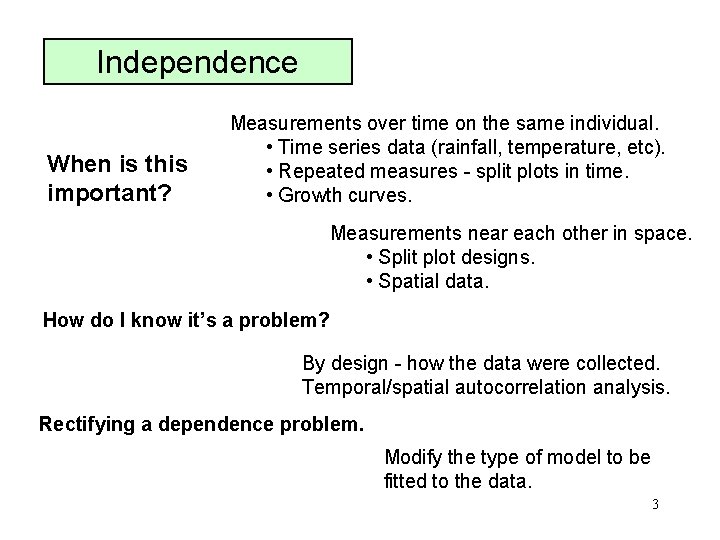
Independence When is this important? Measurements over time on the same individual. • Time series data (rainfall, temperature, etc). • Repeated measures - split plots in time. • Growth curves. Measurements near each other in space. • Split plot designs. • Spatial data. How do I know it’s a problem? By design - how the data were collected. Temporal/spatial autocorrelation analysis. Rectifying a dependence problem. Modify the type of model to be fitted to the data. 3

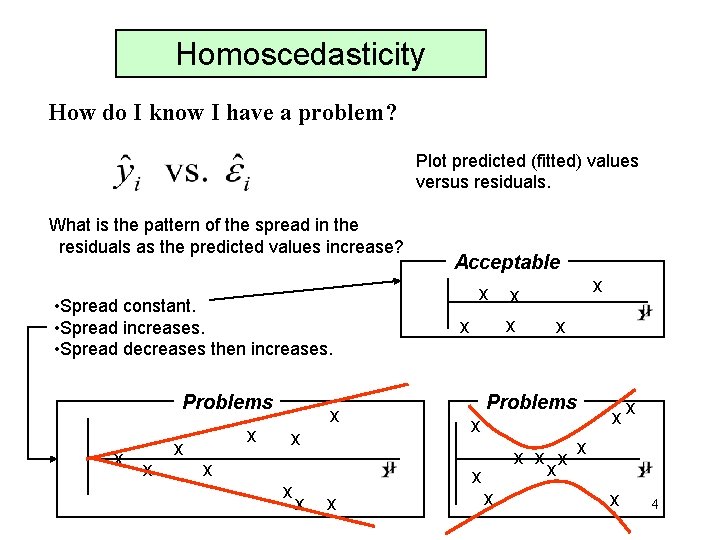
Homoscedasticity How do I know I have a problem? Plot predicted (fitted) values versus residuals. What is the pattern of the spread in the residuals as the predicted values increase? • Spread constant. • Spread increases. • Spread decreases then increases. Problems x x x x Acceptable x x x Problems x x xx x x x 4

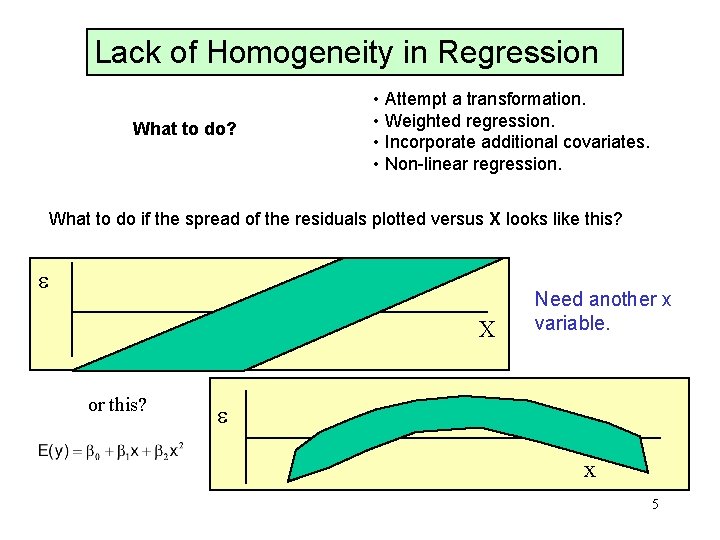
Lack of Homogeneity in Regression What to do? • Attempt a transformation. • Weighted regression. • Incorporate additional covariates. • Non-linear regression. What to do if the spread of the residuals plotted versus X looks like this? e X or this? Need another x variable. e x 5

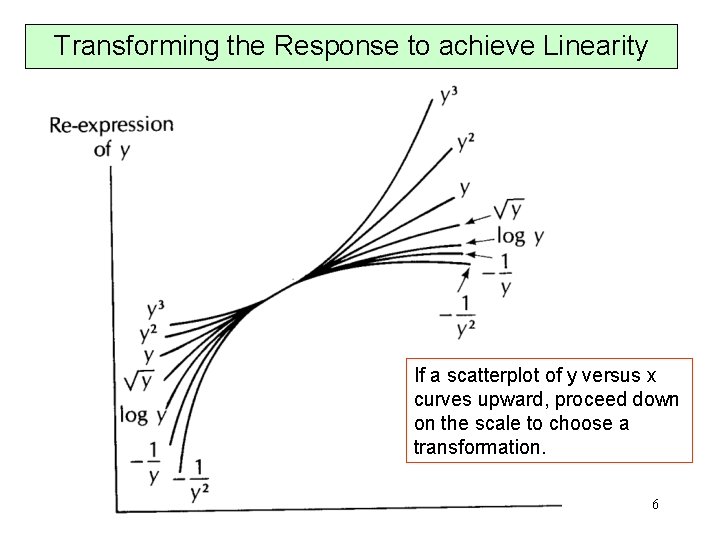
Transforming the Response to achieve Linearity If a scatterplot of y versus x curves upward, proceed down on the scale to choose a transformation. 6

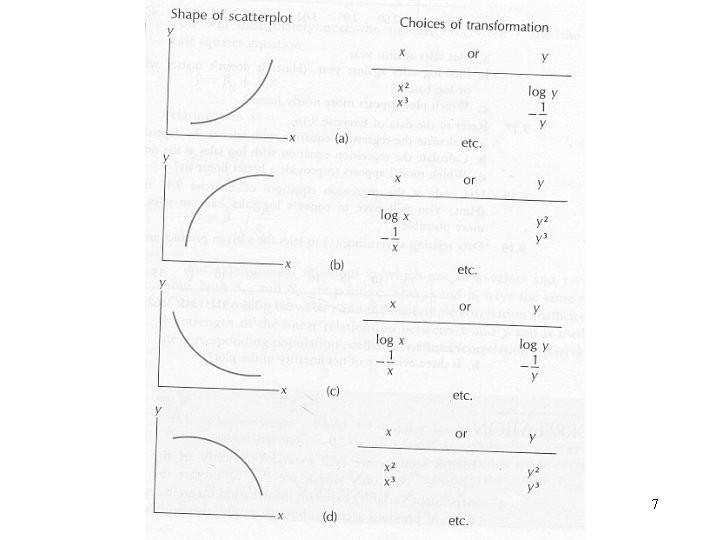
7

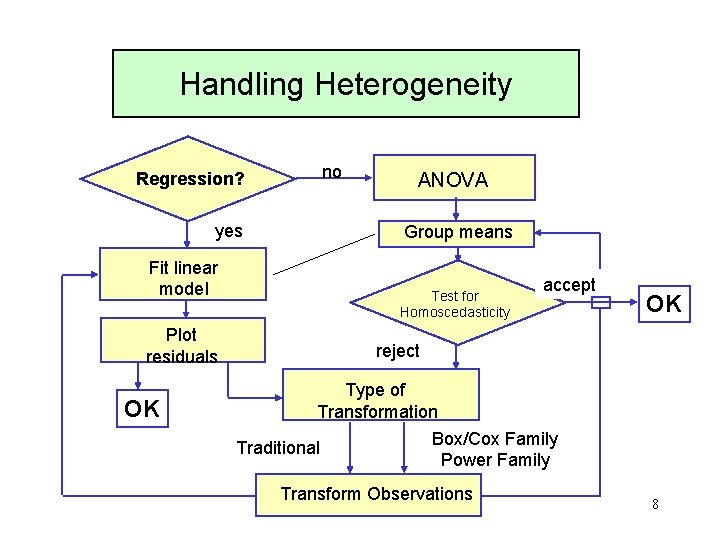
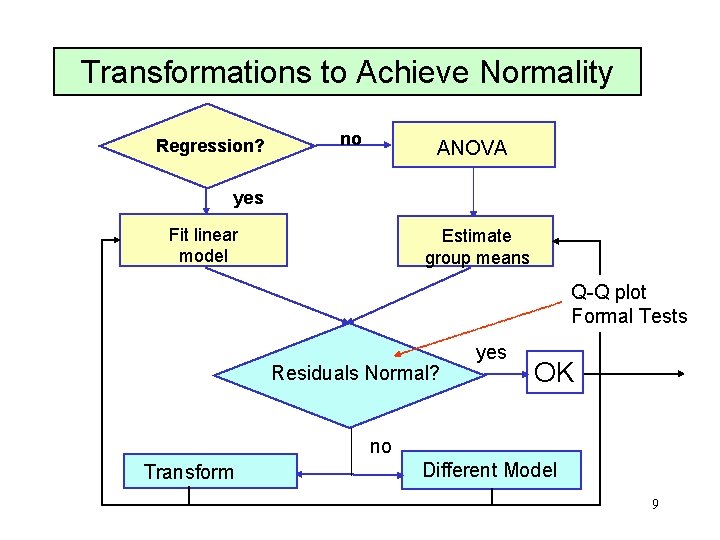
Handling Heterogeneity Regression? yes Fit linear model Plot residuals OK no ANOVA Group means Test for Homoscedasticity accept OK reject Type of Transformation Box/Cox Family Traditional Power Family Transform Observations 8

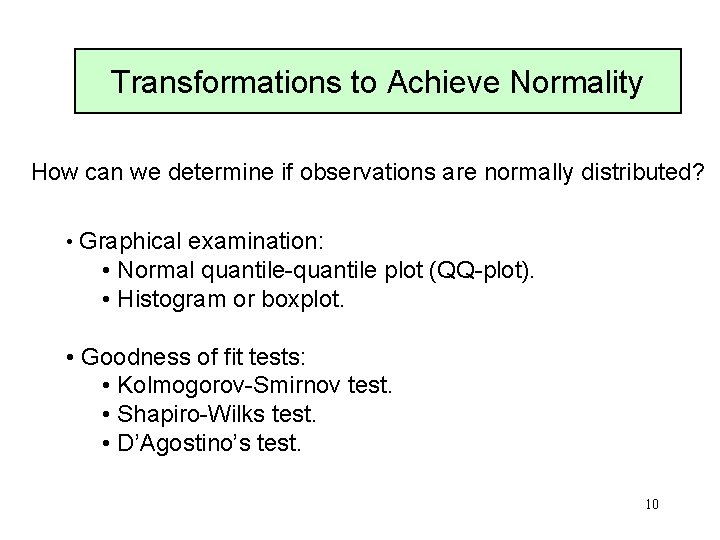
Transformations to Achieve Normality Regression? no ANOVA yes Fit linear model Estimate group means Q-Q plot Formal Tests Residuals Normal? yes OK no Transform Different Model 9

Transformations to Achieve Normality How can we determine if observations are normally distributed? • Graphical examination: • Normal quantile-quantile plot (QQ-plot). • Histogram or boxplot. • Goodness of fit tests: • Kolmogorov-Smirnov test. • Shapiro-Wilks test. • D’Agostino’s test. 10

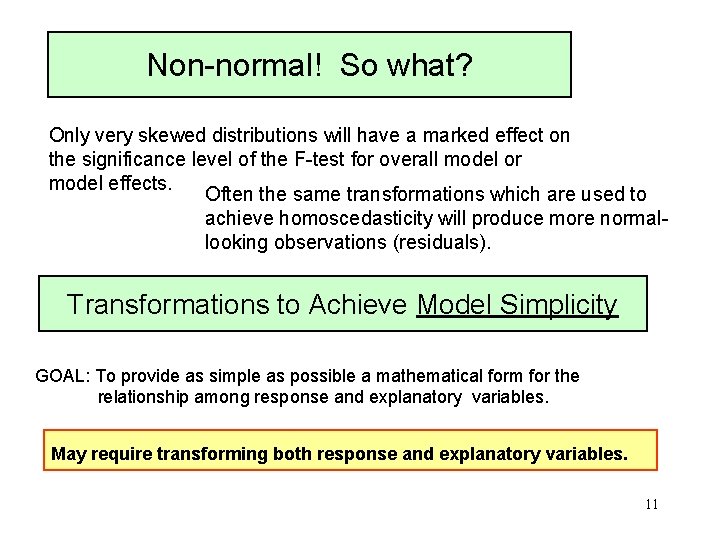
Non-normal! So what? Only very skewed distributions will have a marked effect on the significance level of the F-test for overall model or model effects. Often the same transformations which are used to achieve homoscedasticity will produce more normallooking observations (residuals). Transformations to Achieve Model Simplicity GOAL: To provide as simple as possible a mathematical form for the relationship among response and explanatory variables. May require transforming both response and explanatory variables. 11

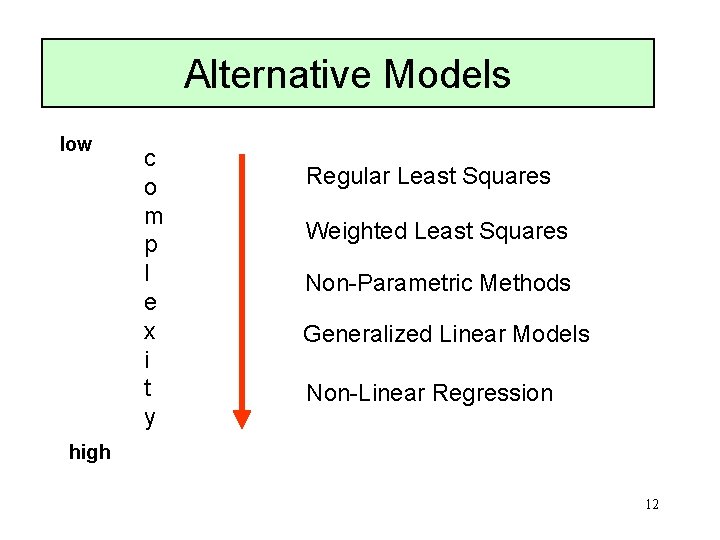
Alternative Models low c o m p l e x i t y Regular Least Squares Weighted Least Squares Non-Parametric Methods Generalized Linear Models Non-Linear Regression high 12

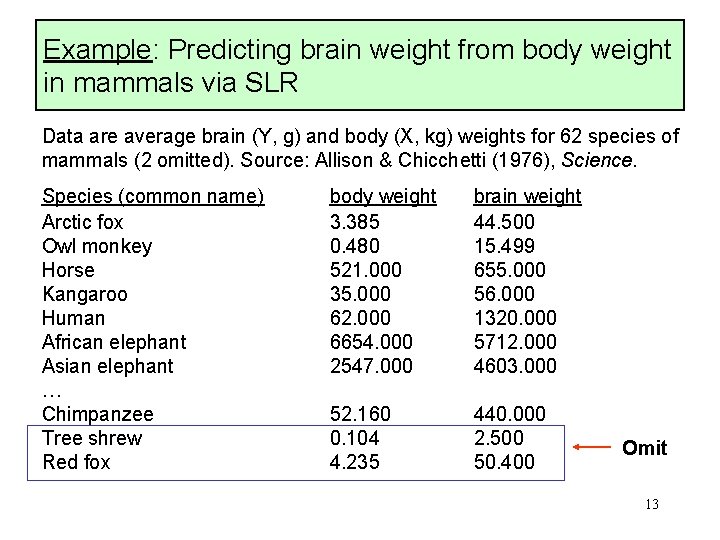
Example: Predicting brain weight from body weight in mammals via SLR Data are average brain (Y, g) and body (X, kg) weights for 62 species of mammals (2 omitted). Source: Allison & Chicchetti (1976), Science. Species (common name) Arctic fox Owl monkey Horse Kangaroo Human African elephant Asian elephant … Chimpanzee Tree shrew Red fox body weight 3. 385 0. 480 521. 000 35. 000 62. 000 6654. 000 2547. 000 brain weight 44. 500 15. 499 655. 000 56. 000 1320. 000 5712. 000 4603. 000 52. 160 0. 104 4. 235 440. 000 2. 500 50. 400 Omit 13

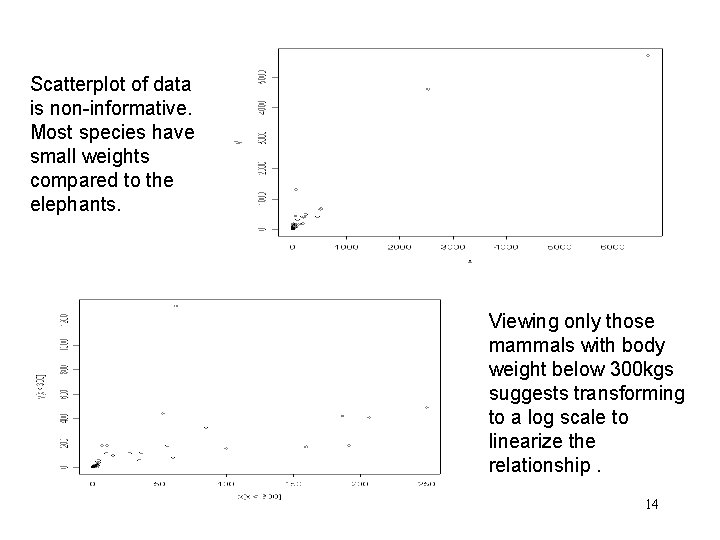
Scatterplot of data is non-informative. Most species have small weights compared to the elephants. Viewing only those mammals with body weight below 300 kgs suggests transforming to a log scale to linearize the relationship. 14

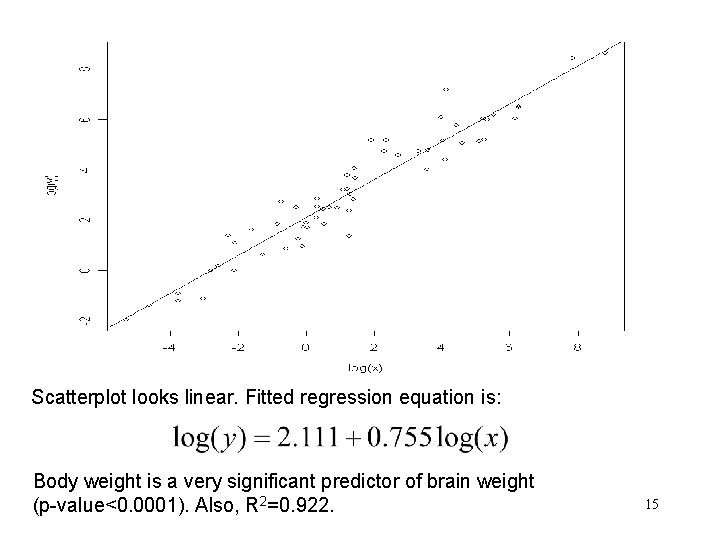
Scatterplot looks linear. Fitted regression equation is: Body weight is a very significant predictor of brain weight (p-value<0. 0001). Also, R 2=0. 922. 15

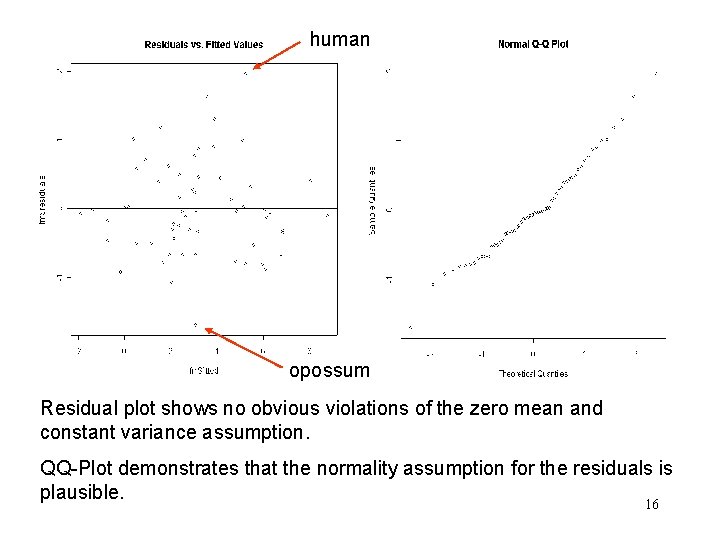
human opossum Residual plot shows no obvious violations of the zero mean and constant variance assumption. QQ-Plot demonstrates that the normality assumption for the residuals is plausible. 16

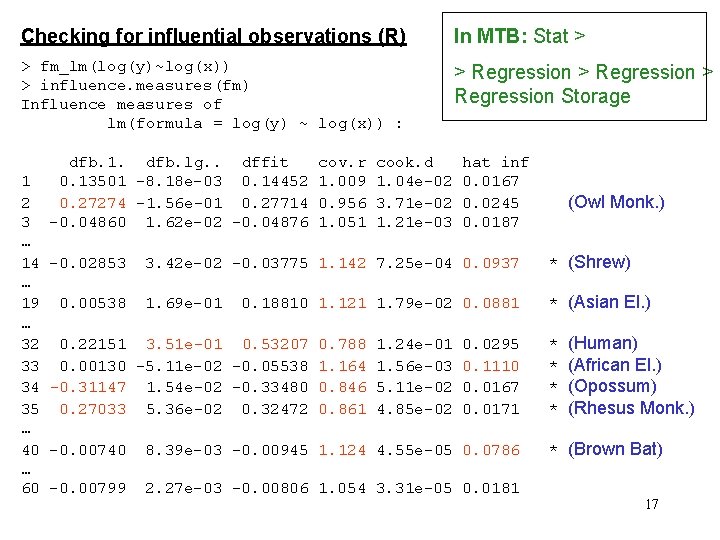
Checking for influential observations (R) In MTB: Stat > > fm_lm(log(y)~log(x)) > influence. measures(fm) Influence measures of lm(formula = log(y) ~ log(x)) : > Regression Storage 1 2 3 … 14 … 19 … 32 33 34 35 … 40 … 60 dfb. 1. dfb. lg. . dffit 0. 13501 -8. 18 e-03 0. 14452 0. 27274 -1. 56 e-01 0. 27714 -0. 04860 1. 62 e-02 -0. 04876 -0. 02853 0. 00538 cov. r 1. 009 0. 956 1. 051 cook. d 1. 04 e-02 3. 71 e-02 1. 21 e-03 hat inf 0. 0167 0. 0245 0. 0187 (Owl Monk. ) 3. 42 e-02 -0. 03775 1. 142 7. 25 e-04 0. 0937 * (Shrew) 1. 69 e-01 * (Asian El. ) 0. 18810 1. 121 1. 79 e-02 0. 0881 0. 22151 3. 51 e-01 0. 53207 0. 788 0. 00130 -5. 11 e-02 -0. 05538 1. 164 -0. 31147 1. 54 e-02 -0. 33480 0. 846 0. 27033 5. 36 e-02 0. 32472 0. 861 1. 24 e-01 1. 56 e-03 5. 11 e-02 4. 85 e-02 0. 0295 0. 1110 0. 0167 0. 0171 -0. 00740 8. 39 e-03 -0. 00945 1. 124 4. 55 e-05 0. 0786 -0. 00799 2. 27 e-03 -0. 00806 1. 054 3. 31 e-05 0. 0181 * * (Human) (African El. ) (Opossum) (Rhesus Monk. ) * (Brown Bat) 17

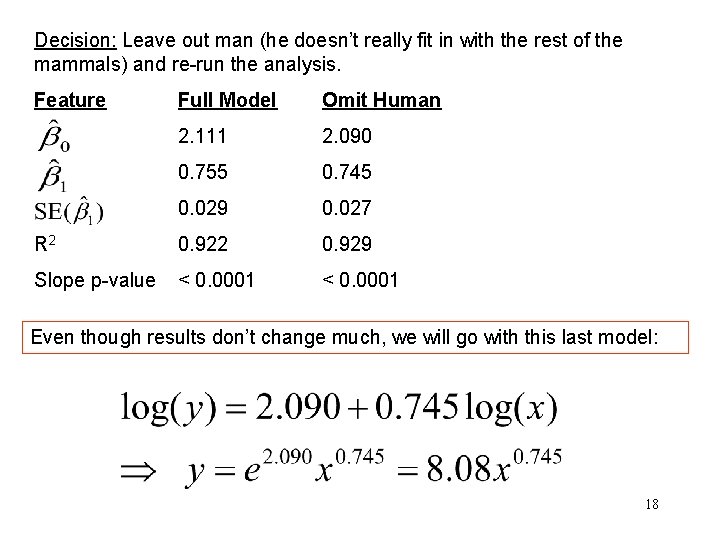
Decision: Leave out man (he doesn’t really fit in with the rest of the mammals) and re-run the analysis. Feature Full Model Omit Human 2. 111 2. 090 0. 755 0. 745 0. 029 0. 027 R 2 0. 929 Slope p-value < 0. 0001 Even though results don’t change much, we will go with this last model: 18

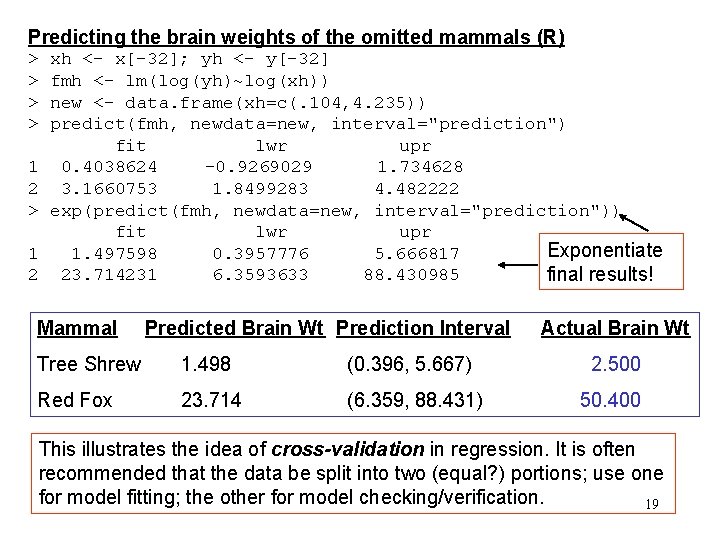
Predicting the brain weights of the omitted mammals (R) > > 1 2 xh <- x[-32]; yh <- y[-32] fmh <- lm(log(yh)~log(xh)) new <- data. frame(xh=c(. 104, 4. 235)) predict(fmh, newdata=new, interval="prediction") fit lwr upr 0. 4038624 -0. 9269029 1. 734628 3. 1660753 1. 8499283 4. 482222 exp(predict(fmh, newdata=new, interval="prediction")) fit lwr upr Exponentiate 1. 497598 0. 3957776 5. 666817 23. 714231 6. 3593633 88. 430985 final results! Mammal Predicted Brain Wt Prediction Interval Actual Brain Wt Tree Shrew 1. 498 (0. 396, 5. 667) 2. 500 Red Fox 23. 714 (6. 359, 88. 431) 50. 400 This illustrates the idea of cross-validation in regression. It is often recommended that the data be split into two (equal? ) portions; use one for model fitting; the other for model checking/verification. 19

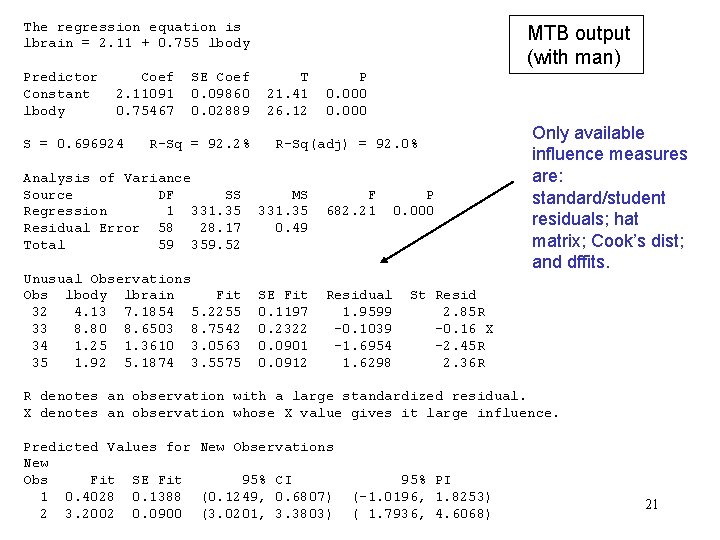
Predicting the brain weights of the omitted mammals (MTB) Influence measures can be selected here. 20

The regression equation is lbrain = 2. 11 + 0. 755 lbody Predictor Constant lbody Coef 2. 11091 0. 75467 S = 0. 696924 SE Coef 0. 09860 0. 02889 R-Sq = 92. 2% Analysis of Variance Source DF SS Regression 1 331. 35 Residual Error 58 28. 17 Total 59 359. 52 Unusual Observations Obs lbody lbrain Fit 32 4. 13 7. 1854 5. 2255 33 8. 80 8. 6503 8. 7542 34 1. 25 1. 3610 3. 0563 35 1. 92 5. 1874 3. 5575 MTB output (with man) T 21. 41 26. 12 P 0. 000 R-Sq(adj) = 92. 0% MS 331. 35 0. 49 F 682. 21 SE Fit 0. 1197 0. 2322 0. 0901 0. 0912 Residual 1. 9599 -0. 1039 -1. 6954 1. 6298 P 0. 000 Only available influence measures are: standard/student residuals; hat matrix; Cook’s dist; and dffits. St Resid 2. 85 R -0. 16 X -2. 45 R 2. 36 R R denotes an observation with a large standardized residual. X denotes an observation whose X value gives it large influence. Predicted Values for New Observations New Obs Fit SE Fit 95% CI 1 0. 4028 0. 1388 (0. 1249, 0. 6807) 2 3. 2002 0. 0900 (3. 0201, 3. 3803) 95% PI (-1. 0196, 1. 8253) ( 1. 7936, 4. 6068) 21