Transcriptomics on Apache Spark Hercules a Map Reduce









- Slides: 29

Transcriptomics on Apache Spark & Hercules a Map. Reduce algorithm for quantifying non-uniform gene expression Cloud. Tech ’ 16 May 24 th-25 th Marrakesh, Morocco Jamie Alnasir & Dr. Hugh Shanahan 25/05/2016

Outline of today’s talk Introduce myself and my research interests Introduce Transcriptomics Quick look over Next-generation sequencing protocol steps that introduce bias Discuss Hercules distributed Map. Reduce algorithm on Apache Spark for assessing Transcriptomic data quality using “bigdata” tools

Jamie Alnasir M. Pharm (Hons) / Ph. D candidate Final year Ph. D research candidate in the department of Computer Science, Royal Holloway University of London Background: Pharmacy and Chemistry (M. Pharm) Kingston University/St. Georges Hospital Medical School, London Currently work at the Centre for Systems and Synthetic Biology (CSSB) - Supervisor Dr. Hugh Shanahan My Ph. D title "The analysis of large biological datasets utilising cloud computing“ My research interests are: The Protein Databank Transcriptomics on Spark (Today’s focus)

Transcriptomics with Apache Spark Today we will look at how a tool used in the Data Science industry can be applied to an important Scientific problem I’ve presented research in Morocco on DNA sequencing and on using Hadoop with The Protein Databank Specifically we will look at applying Apache Spark to the

Introduction to Transcriptomics Transcriptome = the sum total of all the messenger RNA molecules expressed from the genes of an organism Why is the transcriptome of interest? highly dynamic and constantly changing due to intra- and extra-cellular stimuli and disease pathology utilised in biomedical and basic science fields to study… alternative splicing isoforms, post-transcriptional modifications, mutations, gene expression amongst other things… Best way to measure transcriptome variables (gene expression) is using recent Next-generation sequencing technologies (RNA-Seq). This measurement generates “big-data”

Sequencing Data

Problem(s) statement A typical transcriptomics experiment scenario Identifying those transcripts (RNA fragments) whose expression abundance is altered by experimental conditions which differ between sets of samples A good degree of certainty in measurement is crucial A problem… RNA-Seq & Next-gen. sequencing techniques are prone to biases introduced in a number of the steps of a typical sequencing workflow downstream computational methods Transcriptomic sequencing data is produced from an intricate series of chemical reactions that occur during sample preparation, sequencing and data-processing prior to it’s storage……bias can be introduced at these steps…[Gigacience journal published our paper on this] A problem… The data generated by Transcriptomics is vast – “Bigdata”

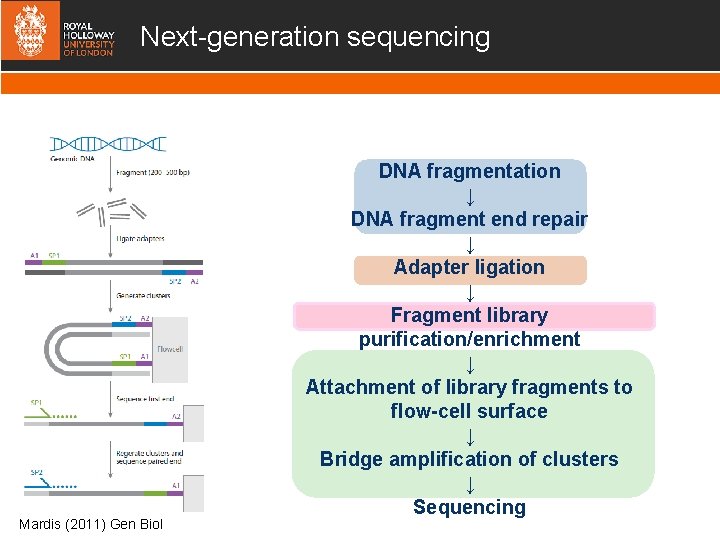
Next-generation sequencing Mardis (2011) Gen Biol DNA fragmentation ↓ DNA fragment end repair ↓ Adapter ligation ↓ Fragment library purification/enrichment ↓ Attachment of library fragments to flow-cell surface ↓ Bridge amplification of clusters ↓ Sequencing

The biases Laboratory steps in which biases were found in the literature Fragmentation DNA fragment end repair Adapter ligation Library enrichment Sequencing process Downstream processing (Basecalling)

Sequencing Data

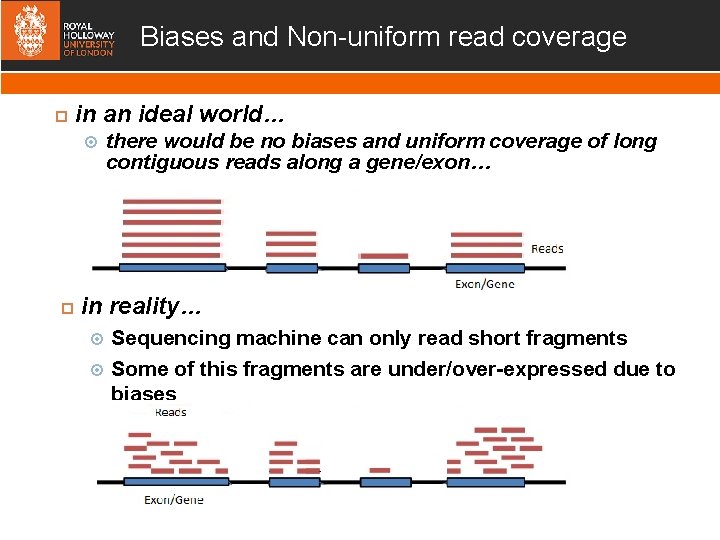
Biases and Non-uniform read coverage in an ideal world… there would be no biases and uniform coverage of long contiguous reads along a gene/exon… in reality… Sequencing machine can only read short fragments Some of this fragments are under/over-expressed due to biases

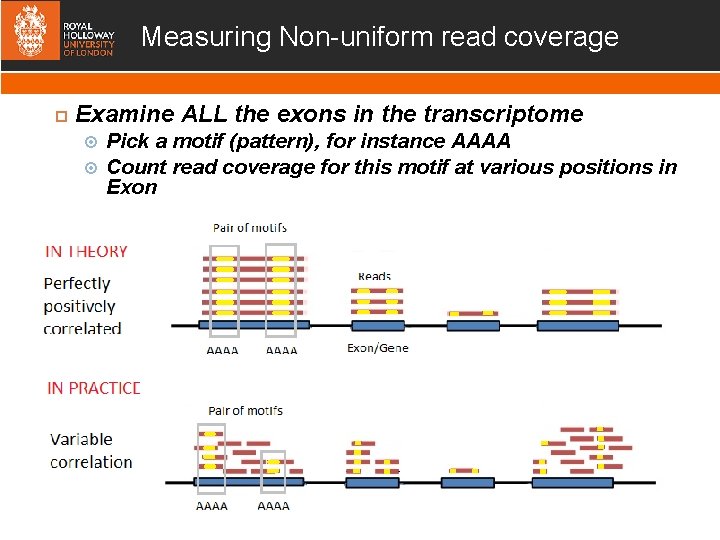
Measuring Non-uniform read coverage Examine ALL the exons in the transcriptome Pick a motif (pattern), for instance AAAA Count read coverage for this motif at various positions in Exon We use Map. Reduce on Apache Spark to do this!

How exactly do we do this?

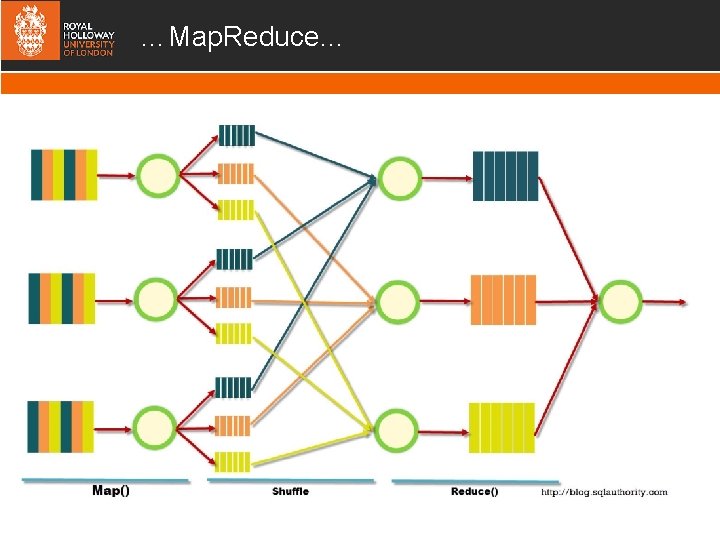
…Map. Reduce…

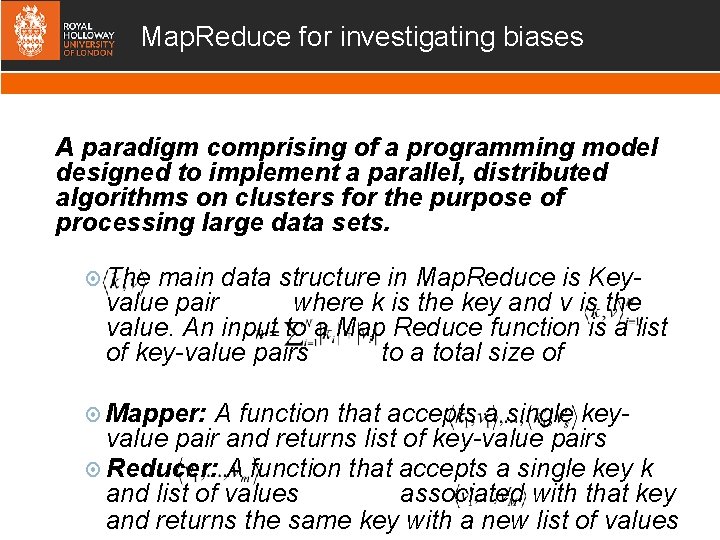
Map. Reduce for investigating biases A paradigm comprising of a programming model designed to implement a parallel, distributed algorithms on clusters for the purpose of processing large data sets. The main data structure in Map. Reduce is Keyvalue pair where k is the key and v is the value. An input to a Map Reduce function is a list of key-value pairs to a total size of Mapper: A function that accepts a single keyvalue pair and returns list of key-value pairs Reducer: A function that accepts a single key k and list of values associated with that key and returns the same key with a new list of values

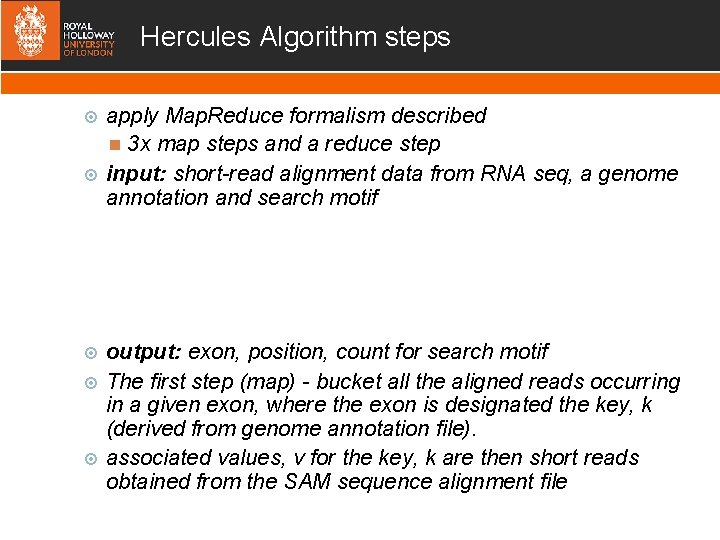
Hercules Algorithm steps apply Map. Reduce formalism described 3 x map steps and a reduce step input: short-read alignment data from RNA seq, a genome annotation and search motif output: exon, position, count for search motif The first step (map) - bucket all the aligned reads occurring in a given exon, where the exon is designated the key, k (derived from genome annotation file). associated values, v for the key, k are then short reads obtained from the SAM sequence alignment file

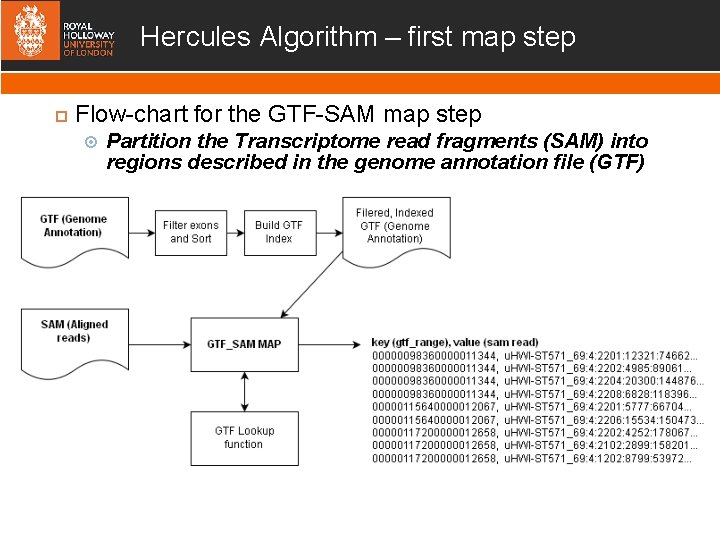
Hercules Algorithm – first map step Flow-chart for the GTF-SAM map step Partition the Transcriptome read fragments (SAM) into regions described in the genome annotation file (GTF)

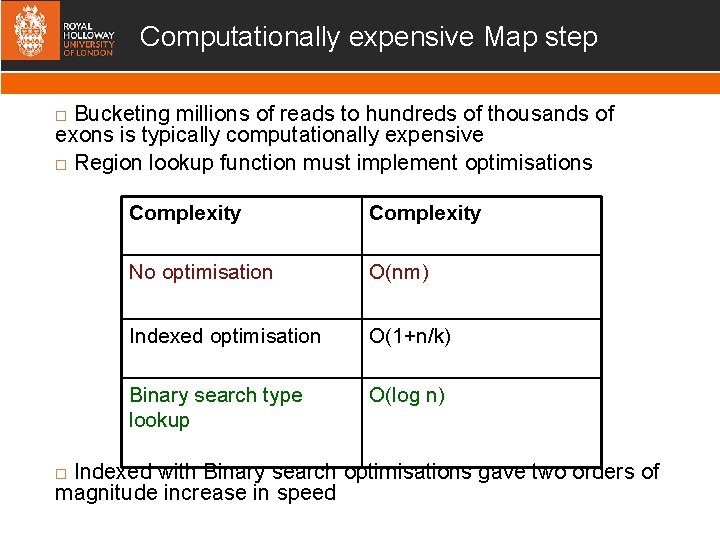
Computationally expensive Map step Bucketing millions of reads to hundreds of thousands of exons is typically computationally expensive Region lookup function must implement optimisations Complexity No optimisation O(nm) Indexed optimisation O(1+n/k) Binary search type lookup O(log n) Indexed with Binary search optimisations gave two orders of magnitude increase in speed

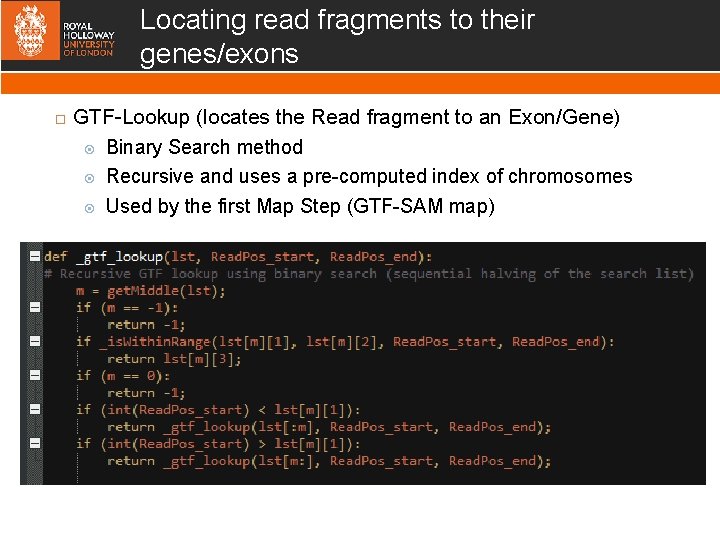
Locating read fragments to their genes/exons GTF-Lookup (locates the Read fragment to an Exon/Gene) Binary Search method Recursive and uses a pre-computed index of chromosomes Used by the first Map Step (GTF-SAM map)

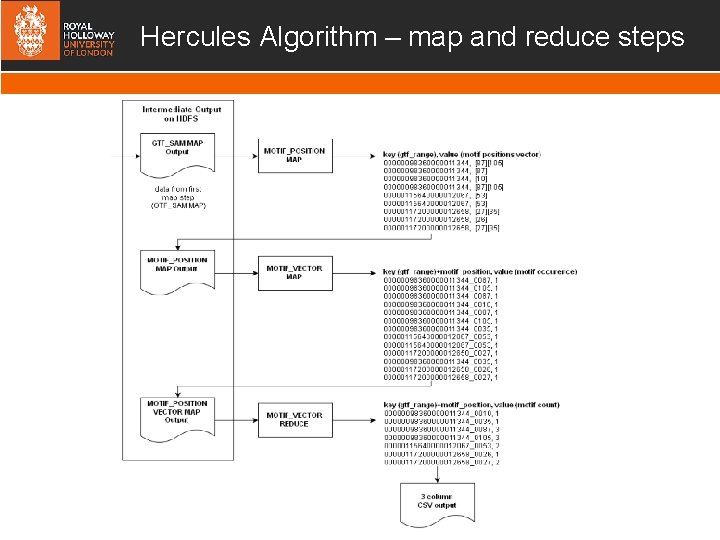
Hercules Algorithm – map and reduce steps

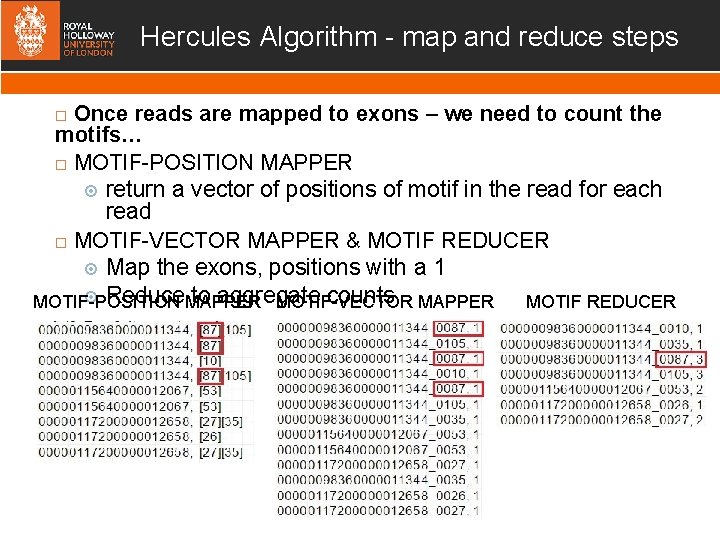
Hercules Algorithm - map and reduce steps Once reads are mapped to exons – we need to count the motifs… MOTIF-POSITION MAPPER return a vector of positions of motif in the read for each read MOTIF-VECTOR MAPPER & MOTIF REDUCER Map the exons, positions with a 1 Reduce MAPPER to aggregate counts MAPPER MOTIF REDUCER MOTIF-POSITION MOTIF-VECTOR

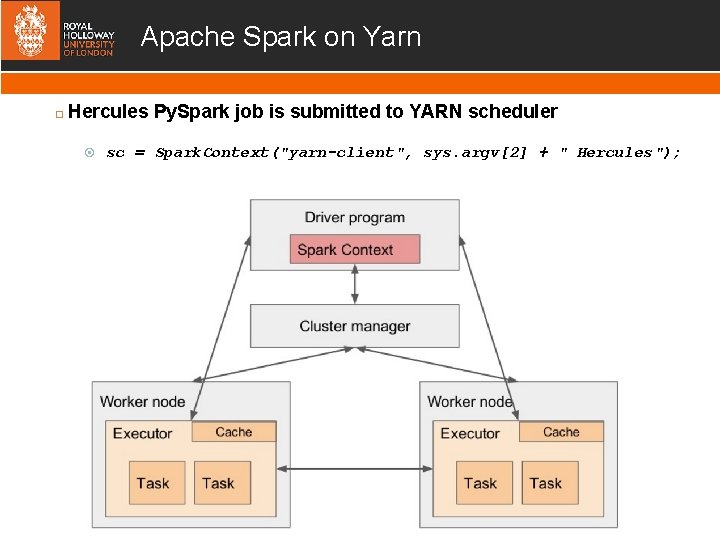
Apache Spark on Yarn Hercules Py. Spark job is submitted to YARN scheduler sc = Spark. Context("yarn-client", sys. argv[2] + " Hercules");

Monitor jobs through Yarn’s cluster Webconsole

How we’ve applied Hercules Algorithm Analysis of biological data – Drosophila Melanogaster transcriptome Analysed all fourmer sequence motifs (i. e. AAAA through GGGG), 44 = 256 daisy-chained Hercules jobs! Interested in counts of fourmers in the same exon at different positions…why? Theory: same motif occurring at different positions in the same exon should have the same counts – i. e. normalised coverage. Literature aware of biases with sequencing of G-C rich regions and repeats We’ve found that not only G-C repeats are difficult/problematic to measure but appears any repeating sequence also can be …lets look at some results…

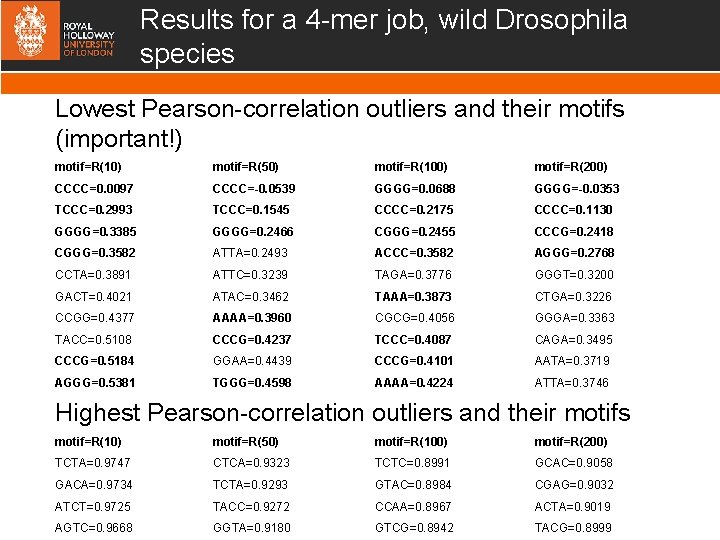
Results for a 4 -mer job, wild Drosophila species Lowest Pearson-correlation outliers and their motifs (important!) motif=R(10) motif=R(50) motif=R(100) motif=R(200) CCCC=0. 0097 CCCC=-0. 0539 GGGG=0. 0688 GGGG=-0. 0353 TCCC=0. 2993 TCCC=0. 1545 CCCC=0. 2175 CCCC=0. 1130 GGGG=0. 3385 GGGG=0. 2466 CGGG=0. 2455 CCCG=0. 2418 CGGG=0. 3582 ATTA=0. 2493 ACCC=0. 3582 AGGG=0. 2768 CCTA=0. 3891 ATTC=0. 3239 TAGA=0. 3776 GGGT=0. 3200 GACT=0. 4021 ATAC=0. 3462 TAAA=0. 3873 CTGA=0. 3226 CCGG=0. 4377 AAAA=0. 3960 CGCG=0. 4056 GGGA=0. 3363 TACC=0. 5108 CCCG=0. 4237 TCCC=0. 4087 CAGA=0. 3495 CCCG=0. 5184 GGAA=0. 4439 CCCG=0. 4101 AATA=0. 3719 AGGG=0. 5381 TGGG=0. 4598 AAAA=0. 4224 ATTA=0. 3746 Highest Pearson-correlation outliers and their motifs motif=R(10) motif=R(50) motif=R(100) motif=R(200) TCTA=0. 9747 CTCA=0. 9323 TCTC=0. 8991 GCAC=0. 9058 GACA=0. 9734 TCTA=0. 9293 GTAC=0. 8984 CGAG=0. 9032 ATCT=0. 9725 TACC=0. 9272 CCAA=0. 8967 ACTA=0. 9019 AGTC=0. 9668 GGTA=0. 9180 GTCG=0. 8942 TACG=0. 8999

What’s next? Incorporate other measures into the algorithm such as GC content Investigate positional biases For instance Hansen et al found the Random Hexamer Priming step in the laboratory can bias tha selection of fragments based on their start/flanking sequences Apply Hercules to compare datasets from the previous study we did on experiment annotation Our paper “Investigation into Annotation of Sequencing steps on the Sequence Read Archive” public database We looked at level of annotation of experiments The question is do poorly annotated transcriptomic experiments show poorer correlation and therefore nonunfiform read coverage?

Thank you!

Thank you! – Any Questions? . . .

References 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. Metzker, M. L. (2010). Sequencing technologies - the next generation. Nature reviews. Genetics, 11(1), 31– 46. doi: 10. 1038/nrg 2626 Mardis, E. R. (2006). Anticipating the 1, 000 dollar genome. Genome Biology, 7(7), 112. doi: 10. 1186/gb-2006 -7 -7 -112 Leinonen, R. , Sugawara, H. , & Shumway, M. (2011). The sequence read archive. Nucleic Acids Research, 39(Database issue), D 19– 21. doi: 10. 1093/nar/gkq 1019 Edgar, R. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30(1), 207– 210. Brazma, A. (2003). Array. Express--a public repository for microarray gene expression data at the EBI. Nucleic Acids Research, 31(1), 68– 71. doi: 10. 1093/nar/gkg 091 Kamps-Hughes, N. , Quimby, A. , Zhu, Z. , & Johnson, E. A. (2013). Massively parallel characterization of restriction endonucleases. Nucleic acids research, 41(11), e 119. Chemistry & Biology, 2004 Keohavong, P. , & Thilly, W. G. (1989). Fidelity of DNA polymerases in DNA amplification. Proceedings of the National Academy of Sciences of the United States of America, 86(23), 9253– 7. Eastberg, J. H. , Pelletier, J. , & Stoddard, B. L. (2004). Recognition of DNA substrates by T 4 bacteriophage polynucleotide kinase. Nucleic acids research, 32(2), 653– 60 Seguin-Orlando, A. , Schubert, M. , Clary, J. , et al (2013). Ligation bias in illumina next-generation DNA libraries: implications for sequencing ancient genomes Acinas, S. G. , et al (2005). PCR-induced sequence artifacts and bias: insights from comparison of two 16 S r. RNA clone libraries constructed from the sample. Applied and Environmental Microbiology, 71(12), 8966– 9. Sikorsky, J. A. , Primerano, D. A. , Fenger, T. W. , & Denvir, J. (2007). DNA damage reduces Taq DNA polymerase fidelity and PCR amplification efficiency. Biochemical and biophysical research communications, 355(2), 431– 7. Meacham et al (2011) Identification and correction of systematic error in high-throughput sequence data - 1471 -2105 -12 -451. pdf. (2011)