Transcriptome Analysis Microarray Technology and Data Analysis Roy




- Slides: 48

Transcriptome Analysis Microarray Technology and Data Analysis Roy Williams Ph. D Sanford | Burnham Medical Research Institute

Microarray Revolution

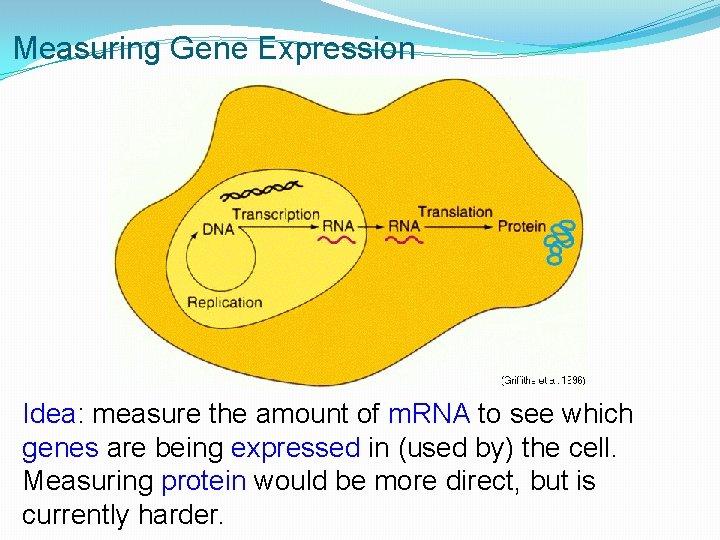
Measuring Gene Expression Idea: measure the amount of m. RNA to see which genes are being expressed in (used by) the cell. Measuring protein would be more direct, but is currently harder.

General assumption of microarray technology �Use m. RNA transcript abundance level as a measure of expression for the corresponding gene �Proportional to degree of gene expression

How to measure RNA abundance �Several different approaches with similar themes �Illumina bead array – highly redundant oligo array �Affymetrix Gene. Chip – highly redundant oligo array �Nimblegen – highly redundant long oligo array � 2 -colour array (very long c. DNA; low redundancy) �SAGE (random Sanger sequencing of c. DNA library) �Reborn as Next Gen RNA seq

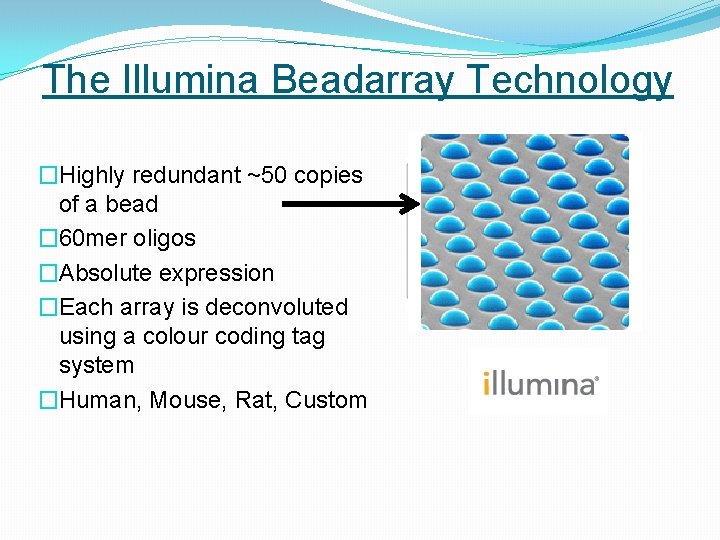
The Illumina Beadarray Technology �Highly redundant ~50 copies of a bead � 60 mer oligos �Absolute expression �Each array is deconvoluted using a colour coding tag system �Human, Mouse, Rat, Custom

Affymetrix Technology �Highly redundant (~25 short oligos per gene) �Absolute expression �PM-MM oligo system valuable for cross hybe detection �Human, Mouse, E. coli, Yeast……. . �Affy and illumina arrays have been systematically compared

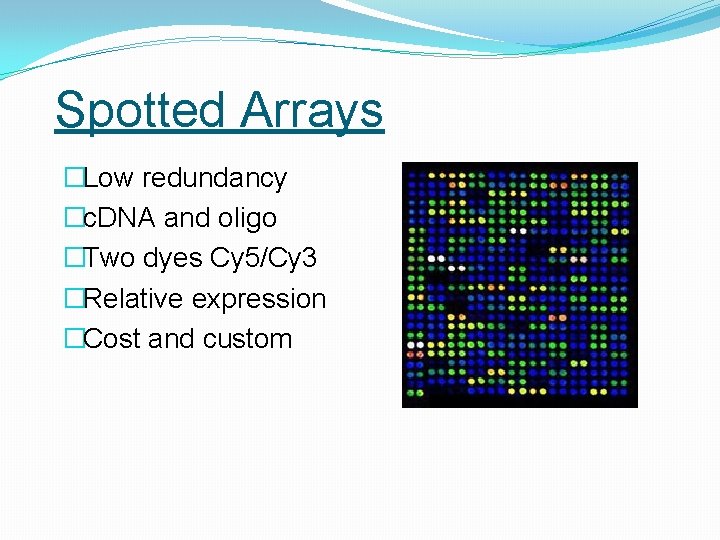
Spotted Arrays �Low redundancy �c. DNA and oligo �Two dyes Cy 5/Cy 3 �Relative expression �Cost and custom

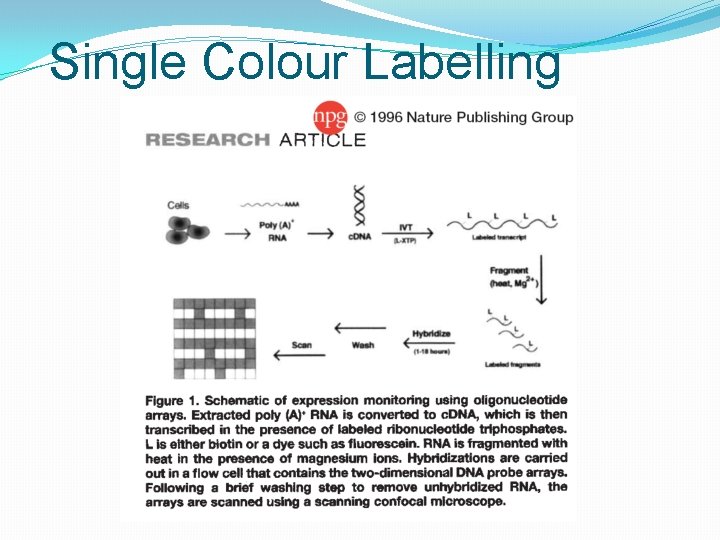
Single Colour Labelling

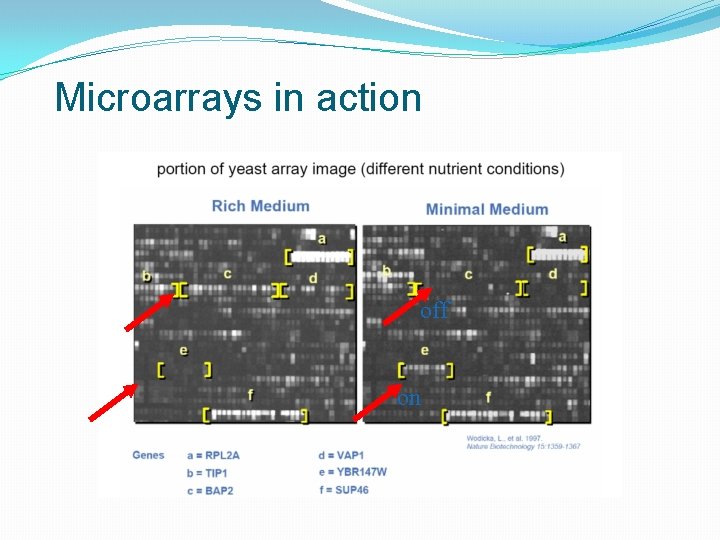
Microarrays in action off on

Areas Being Studied with Microarrays �Differential gene expression between two (or more) sample types �Similar gene expression across treatments �Tumour sub-class identification using gene expression profiles �Classification of malignancies into known classes �Identification of “marker” genes that characterize different cell types �Identification of genes associated with clinical outcomes (e. g. survival)

Experimental Design Experiment Perform Experiment • Replicates • 2 x 3 chips • <2 x 5 chips • Standardize conditions • Dump outliers

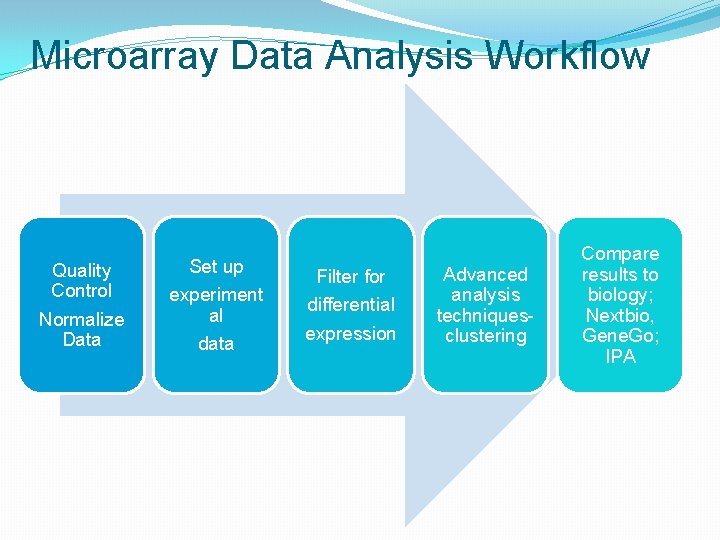
Microarray Data Analysis Workflow Quality Control Normalize Data Set up experiment al data Filter for differential expression Advanced analysis techniquesclustering Compare results to biology; Nextbio, Gene. Go; IPA

Recommended Software �Free Software – Gene. Pattern -- powerful, many plug-in packages and pipelines -- good video examples/tutorials �Gene. Spring GX 11 �R-Bioconductor (with guidance) �Hierarchical Cluster Explorer – easy clustering �Cytoscape, GSEA – for pathway visualisation �Partek �IPA, Nextbio, Gene. Go <= Burnham subscriptions!

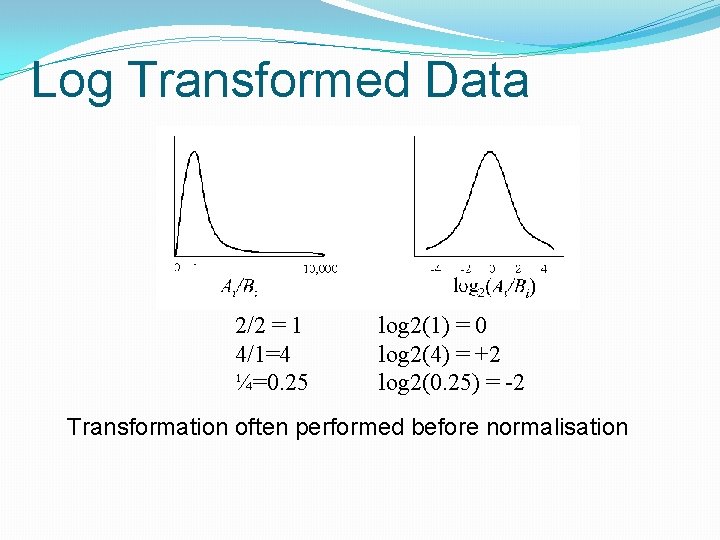
Log Transformed Data 2/2 = 1 4/1=4 ¼=0. 25 log 2(1) = 0 log 2(4) = +2 log 2(0. 25) = -2 Transformation often performed before normalisation

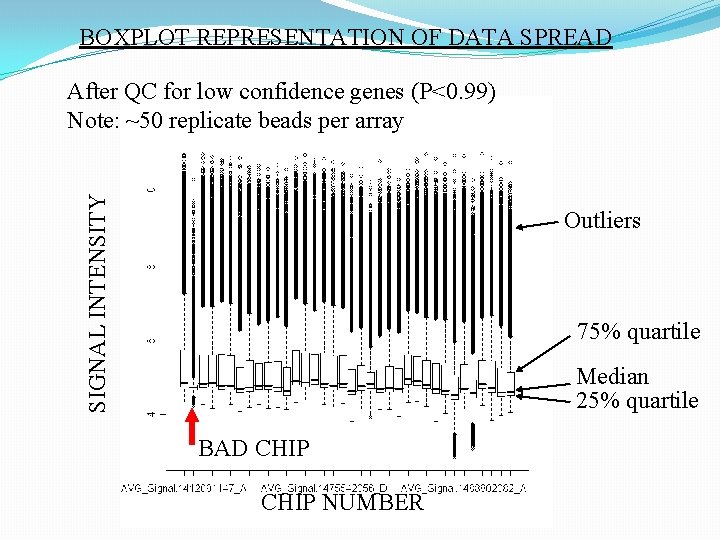
BOXPLOT REPRESENTATION OF DATA SPREAD SIGNAL INTENSITY After QC for low confidence genes (P<0. 99) Note: ~50 replicate beads per array Outliers 75% quartile Median 25% quartile BAD CHIP NUMBER

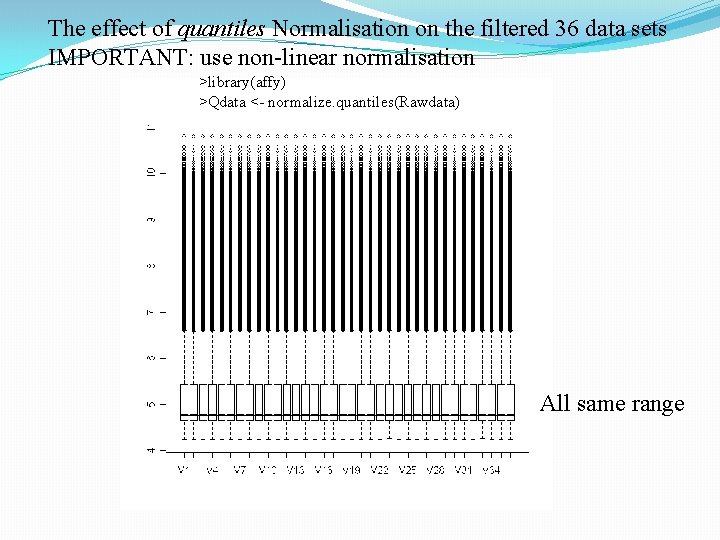
The effect of quantiles Normalisation on the filtered 36 data sets IMPORTANT: use non-linear normalisation >library(affy) >Qdata <- normalize. quantiles(Rawdata) All same range

Data Analysis Examples � 1# Illumina arrays with Gene. Spring GX 11 � 2# Affymetrix data, with a Gene. Pattern module �Import, Quality Control, normalize �Detect differentially expressed genes �Pathway analysis

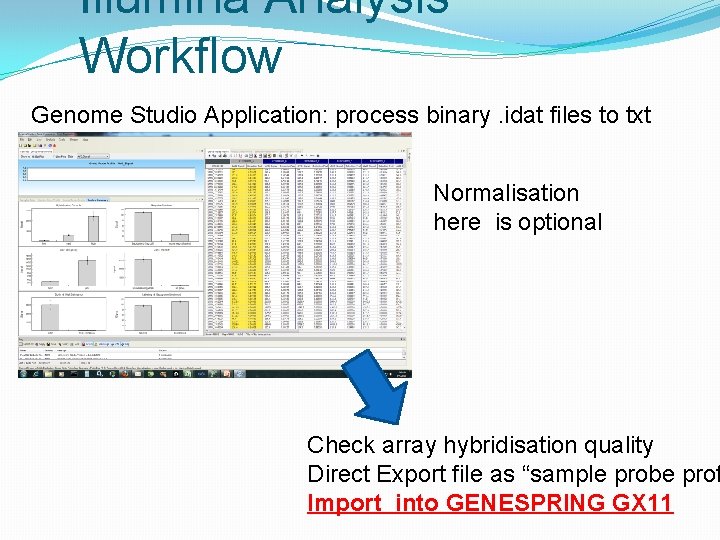
Illumina Analysis Workflow Genome Studio Application: process binary. idat files to txt Normalisation here is optional Check array hybridisation quality Direct Export file as “sample probe prof Import into GENESPRING GX 11

Gene. Spring GX 11 features �Guided workflows �Pathways �GSEA �IPA integration �Ontologies �My. SQL �R script API

Gene. Spring GX 11 �Create New Project �Browse to and load Data �Automated install of Genome. Def from Agilent repository

Illumina Advanced Workflow

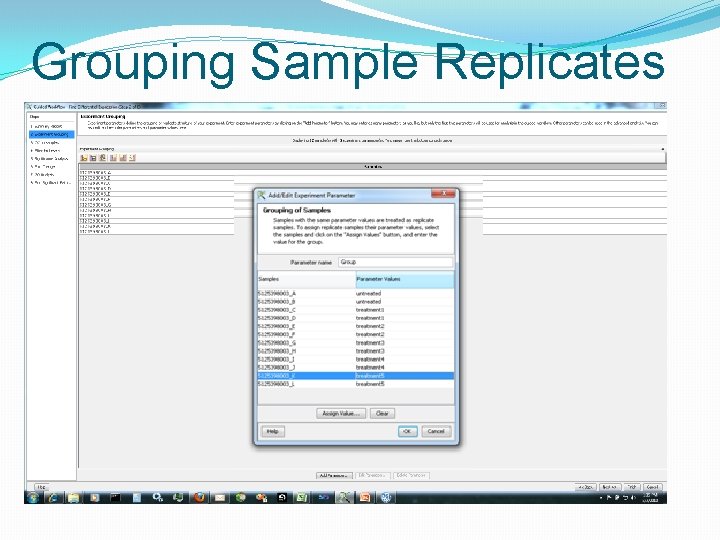
Grouping Sample Replicates

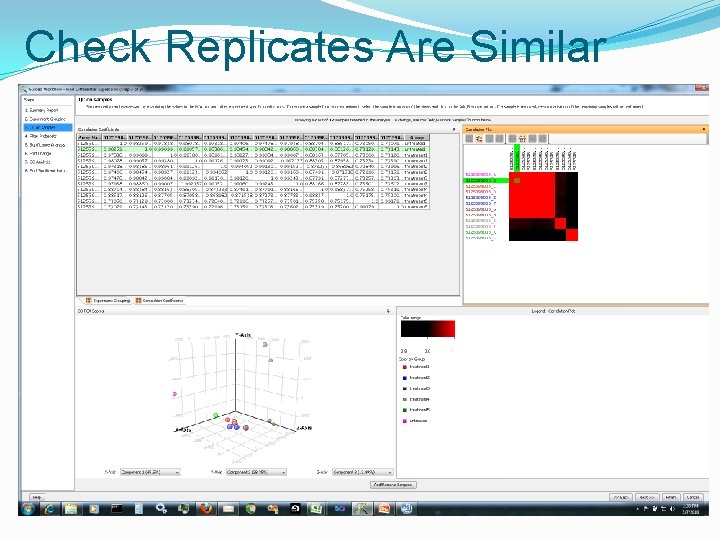
Check Replicates Are Similar

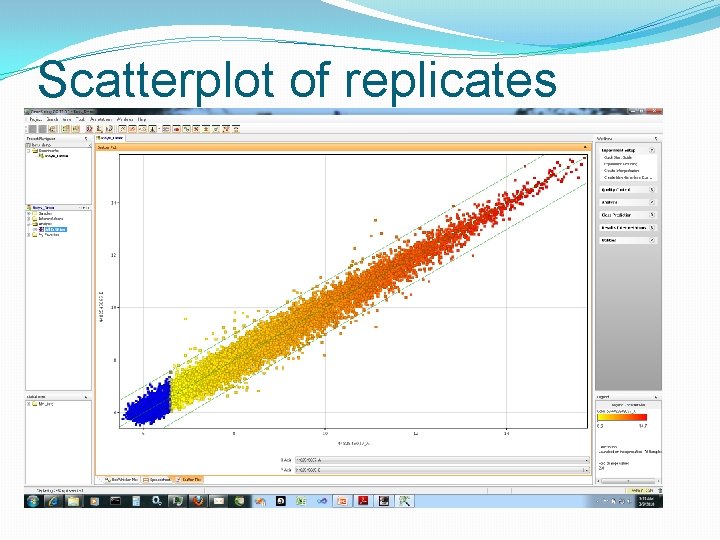
Scatterplot of replicates

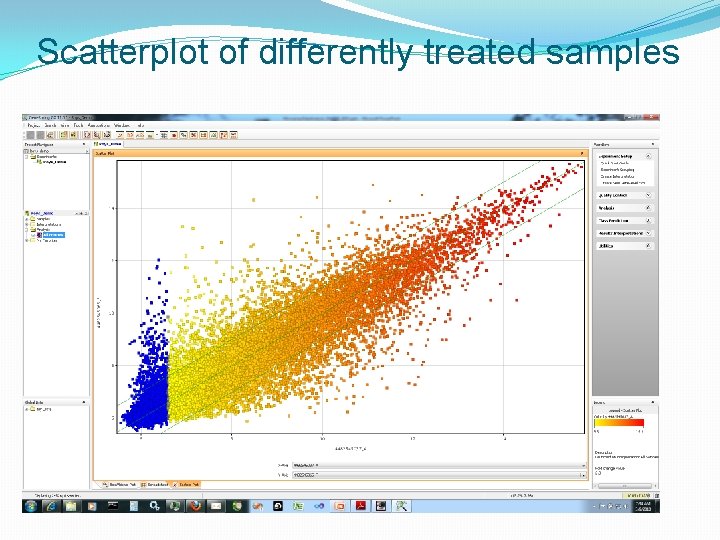
Scatterplot of differently treated samples

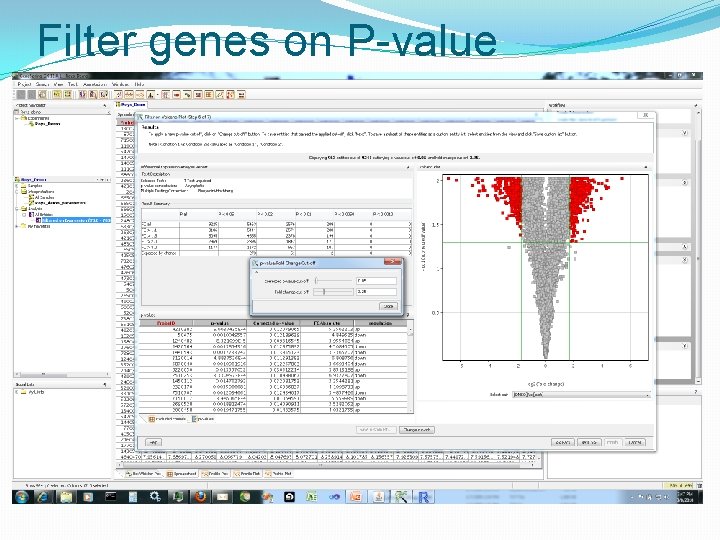
Filter genes on P-value

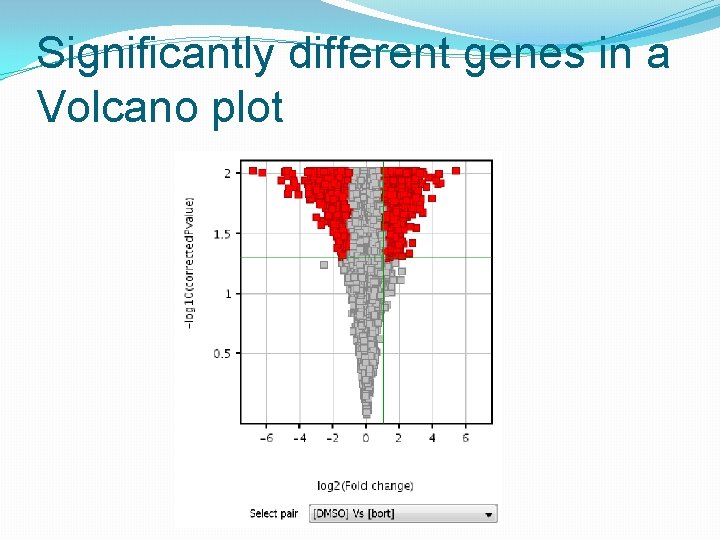
Significantly different genes in a Volcano plot

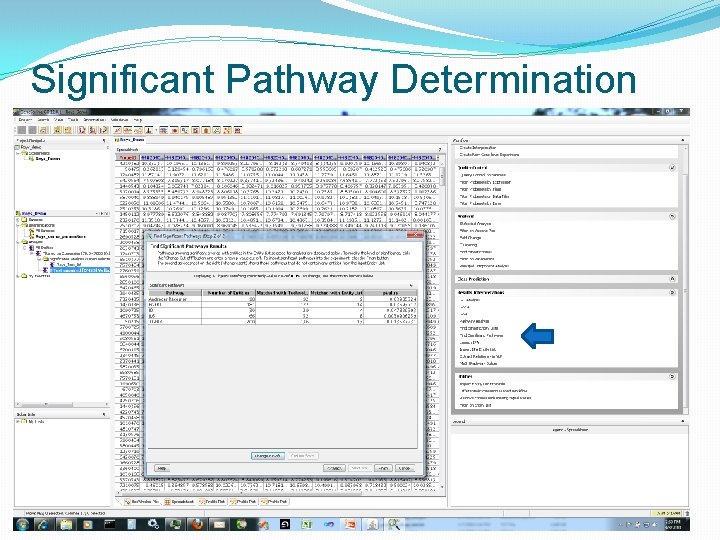
Significant Pathway Determination

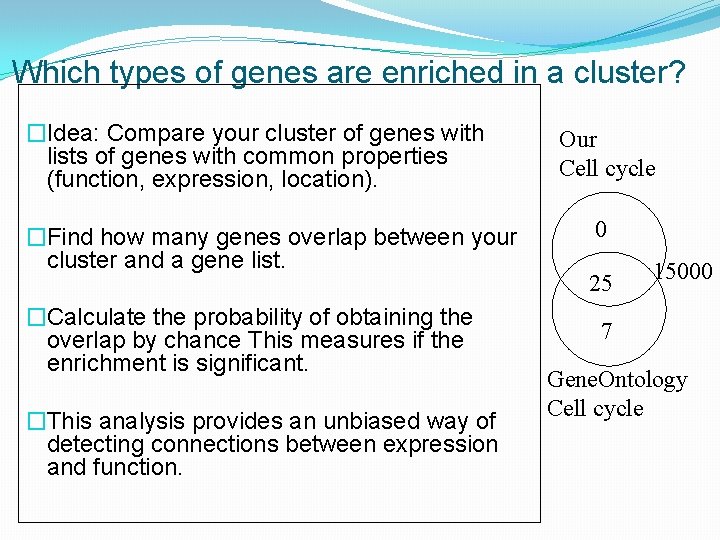
Which types of genes are enriched in a cluster? �Idea: Compare your cluster of genes with lists of genes with common properties (function, expression, location). �Find how many genes overlap between your cluster and a gene list. �Calculate the probability of obtaining the overlap by chance This measures if the enrichment is significant. �This analysis provides an unbiased way of detecting connections between expression and function. Our Cell cycle 0 25 15000 7 Gene. Ontology Cell cycle

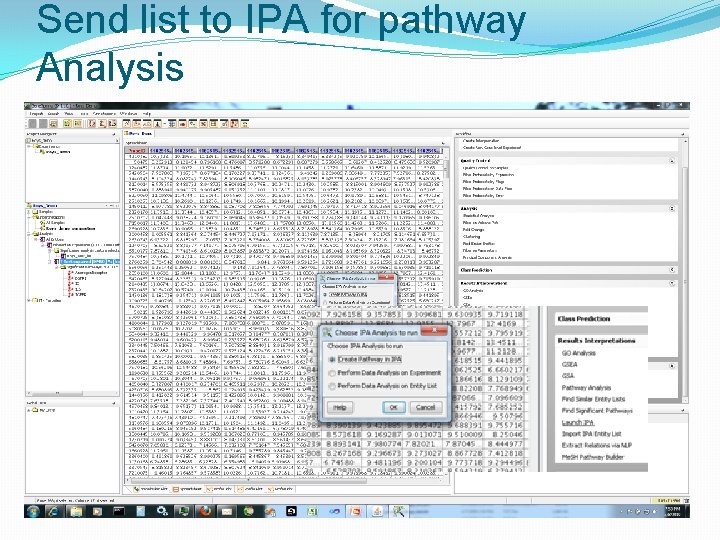
Send list to IPA for pathway Analysis

Significant Pathways sent to Ingenuity Pathway Analysis

Completed Analysis Data genelists Pathways

Affymetrix Workflow: Gene. Pattern

Comparative Marker Selection

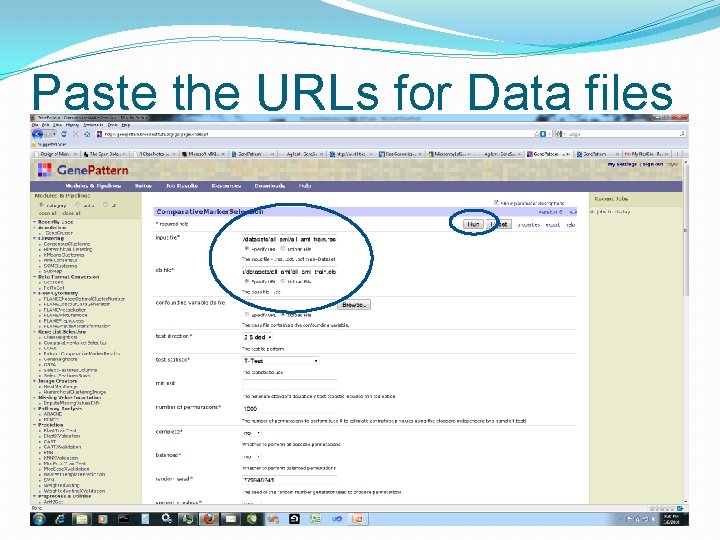
Paste the URLs for Data files

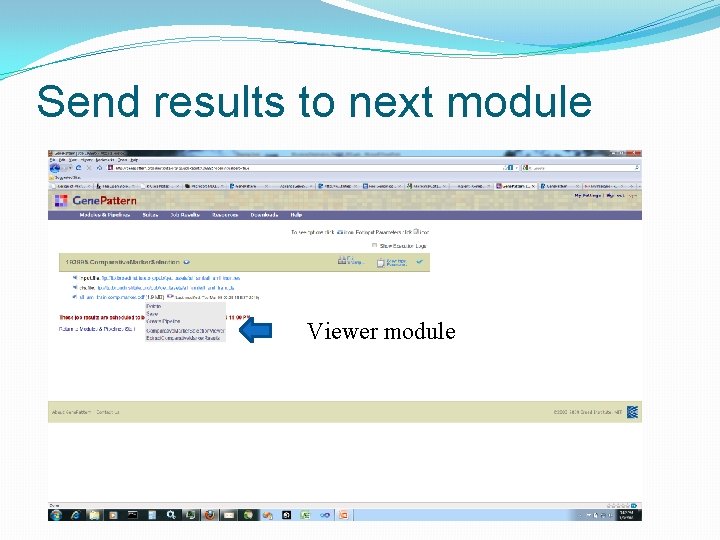
Send results to next module Viewer module

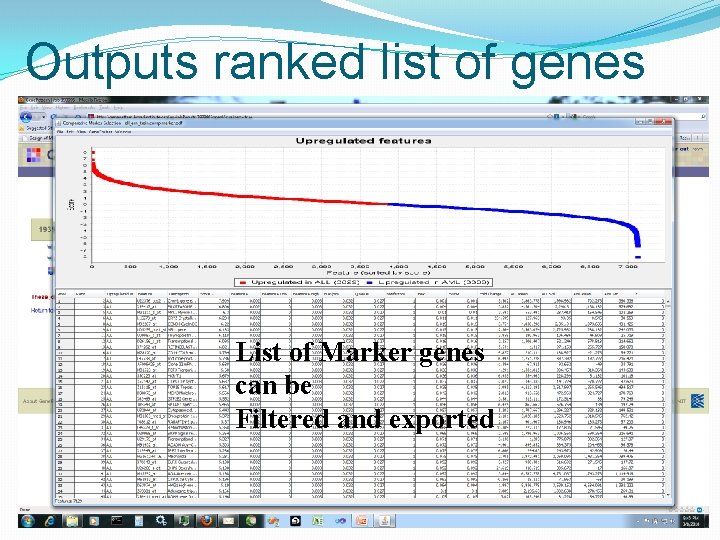
Outputs ranked list of genes List of Marker genes can be Filtered and exported

Nextbio �Compares your Genelists to the Nextbio database �Can reveal unexpected similarities between datasets �Has a very good literature database connected to the results �Contains data from model organisms

Ingenuity Pathway Analysis �Detects networks in your data �Allows you to look for connections between genes and drugs/small molecules �Focused on Man and Mouse Gene. Go High Quality hand annotated ontologies �Has a very good literature database connected to the results �Contains data from model organisms

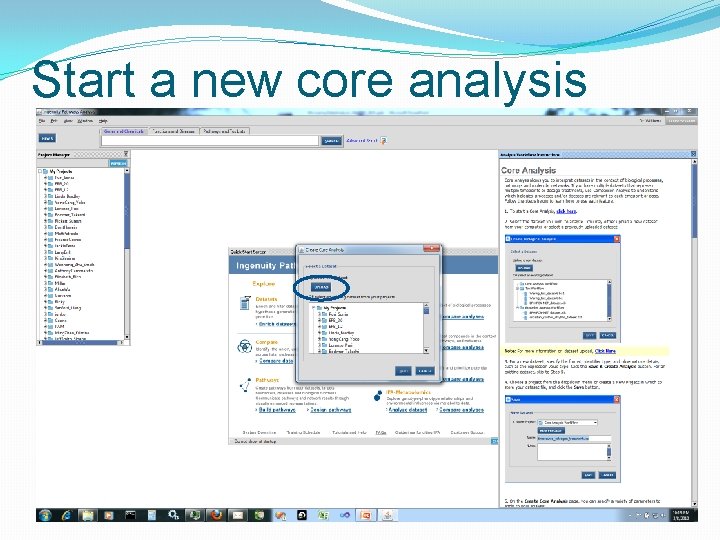
Start a new core analysis

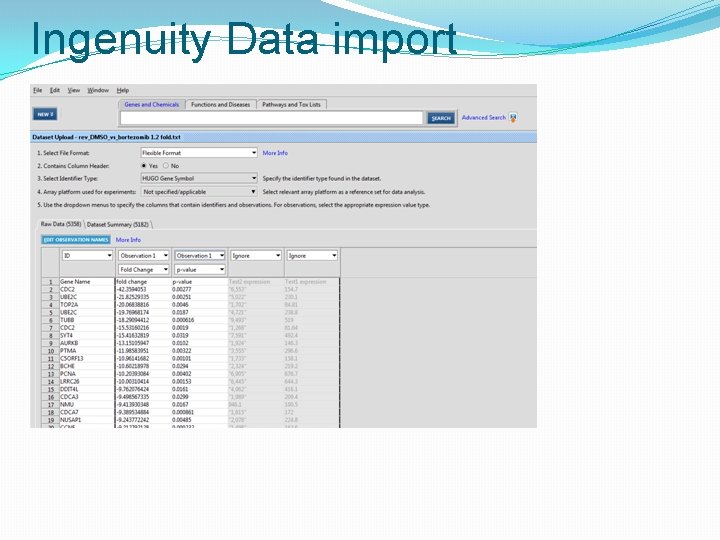
Ingenuity Data import

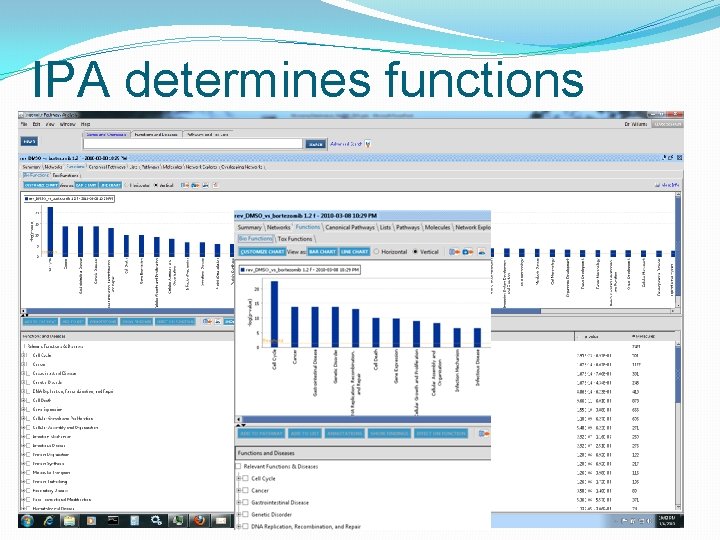
IPA determines functions

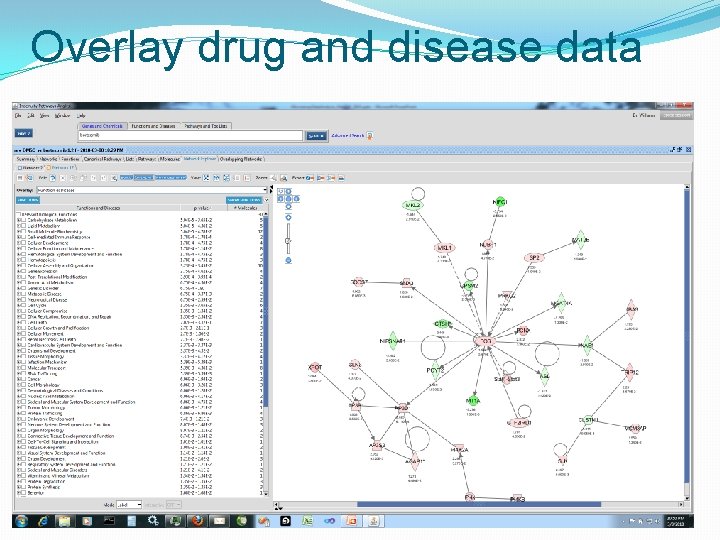
Overlay drug and disease data

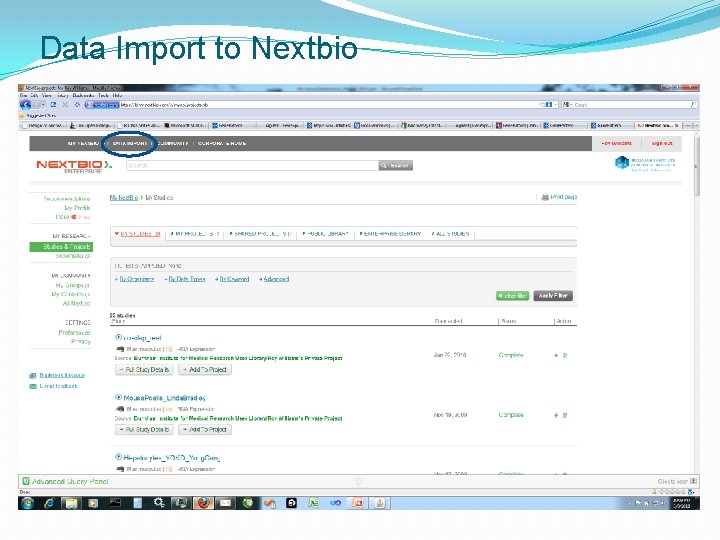
Data Import to Nextbio

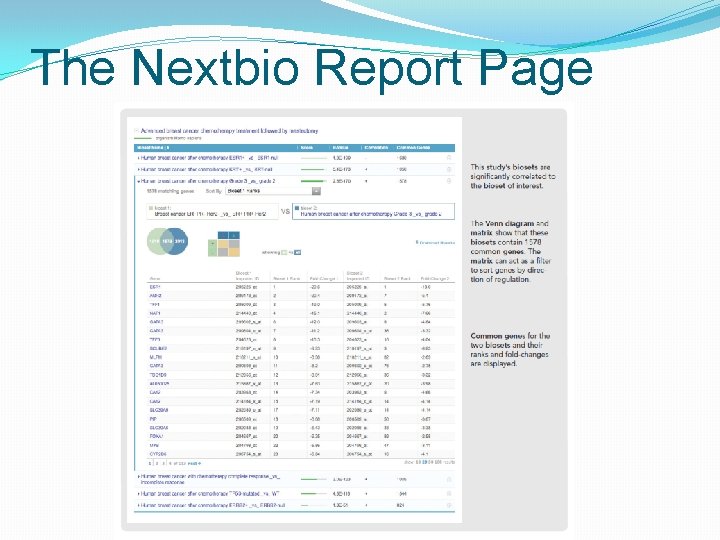
The Nextbio Report Page

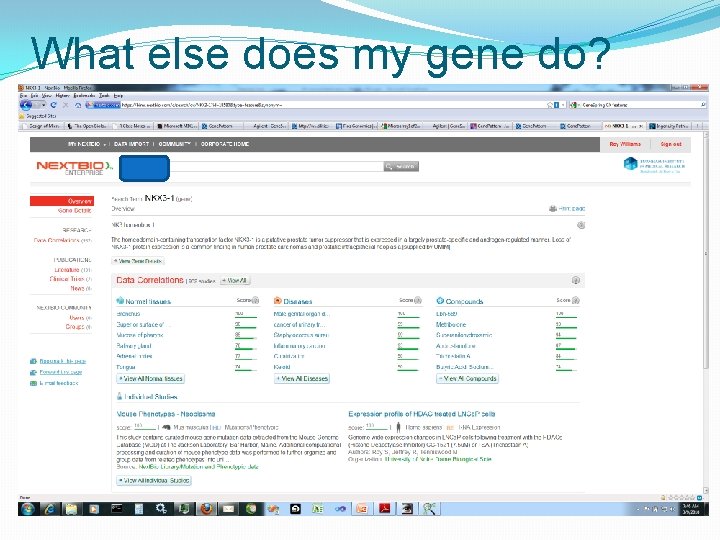
What else does my gene do?

THE END �Many thanks for coming!