Transcription RNA n Structure Ribose both 2 and

Transcription
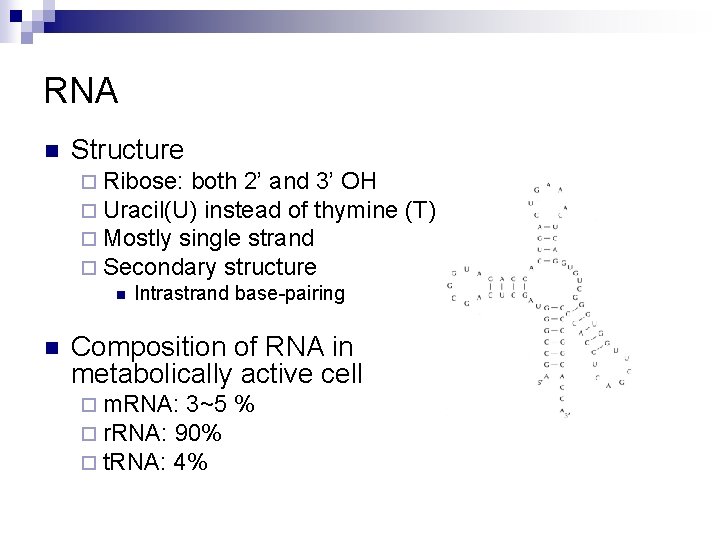
RNA n Structure ¨ Ribose: both 2’ and 3’ OH ¨ Uracil(U) instead of thymine ¨ Mostly single strand ¨ Secondary structure n Intrastrand base-pairing n Composition of RNA in metabolically active cell ¨ m. RNA: 3~5 ¨ r. RNA: 90% ¨ t. RNA: 4% % (T)
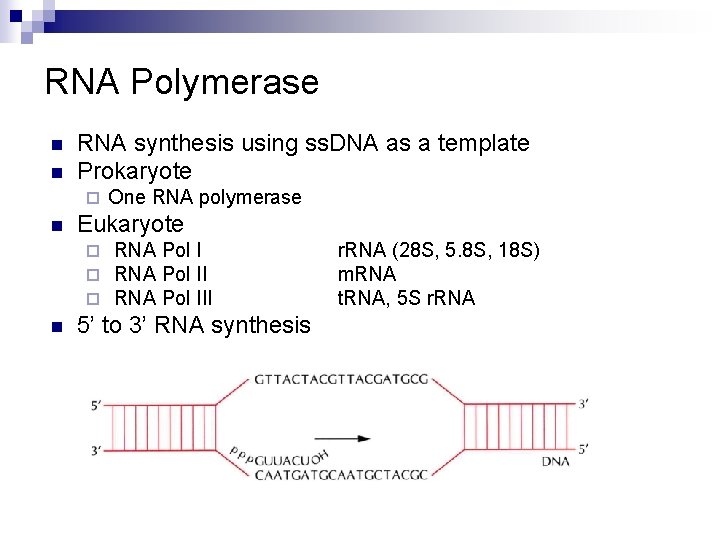
RNA Polymerase n n RNA synthesis using ss. DNA as a template Prokaryote ¨ n Eukaryote ¨ ¨ ¨ n One RNA polymerase RNA Pol III 5’ to 3’ RNA synthesis r. RNA (28 S, 5. 8 S, 18 S) m. RNA t. RNA, 5 S r. RNA
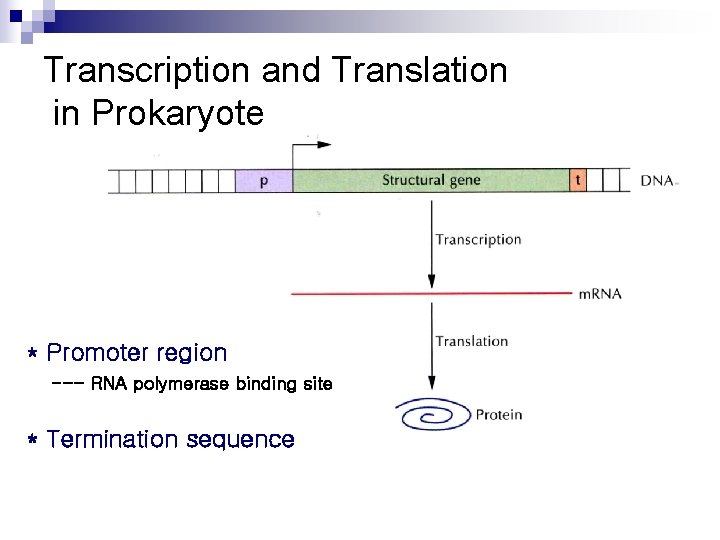
Transcription and Translation in Prokaryote * Promoter region --- RNA polymerase binding site * Termination sequence
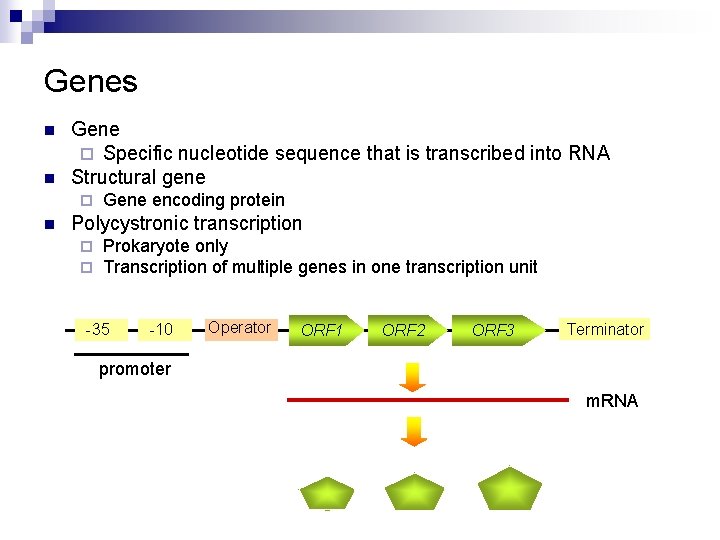
Genes n n Gene ¨ Specific nucleotide sequence that is transcribed into RNA Structural gene ¨ n Gene encoding protein Polycystronic transcription ¨ ¨ Prokaryote only Transcription of multiple genes in one transcription unit -35 -10 Operator ORF 1 ORF 2 ORF 3 Terminator promoter m. RNA
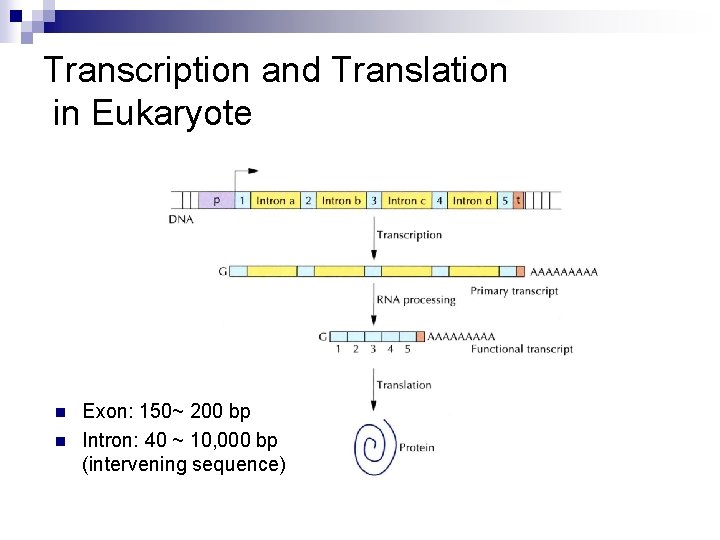
Transcription and Translation in Eukaryote n n Exon: 150~ 200 bp Intron: 40 ~ 10, 000 bp (intervening sequence)
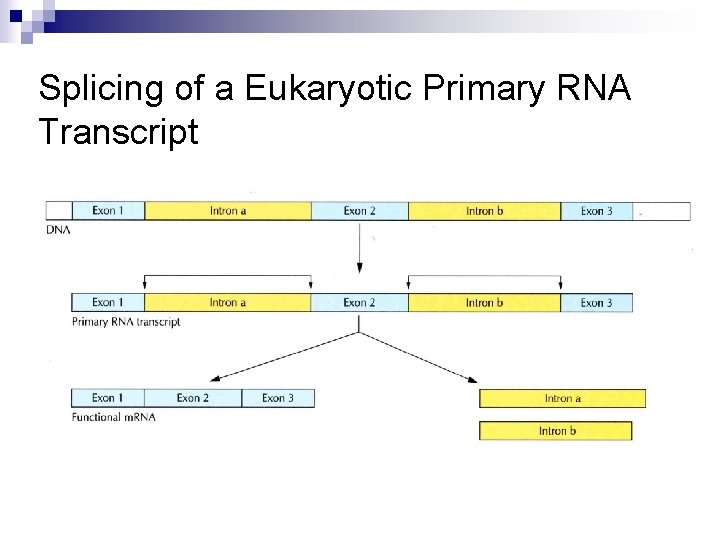
Splicing of a Eukaryotic Primary RNA Transcript
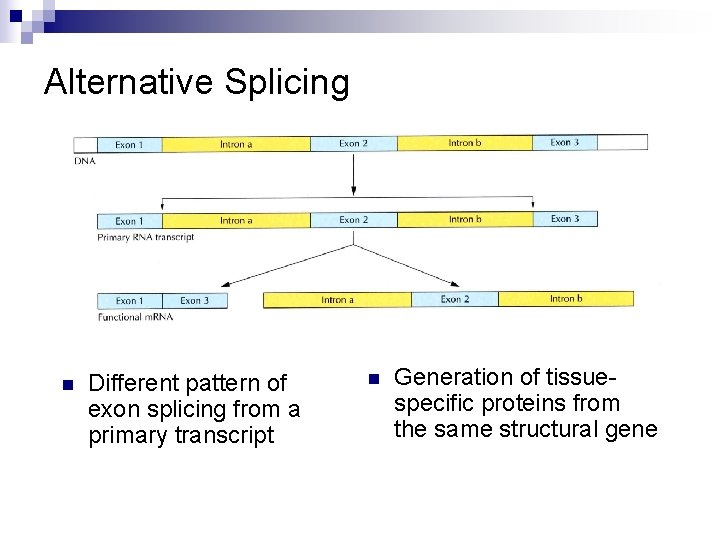
Alternative Splicing n Different pattern of exon splicing from a primary transcript n Generation of tissuespecific proteins from the same structural gene

n A small number of eukaryotic structural genes lack intron (e. g. olfactory genes). n Exon-skipping mechanism generates different gene products in different tissues from the same structural gene.

Translation
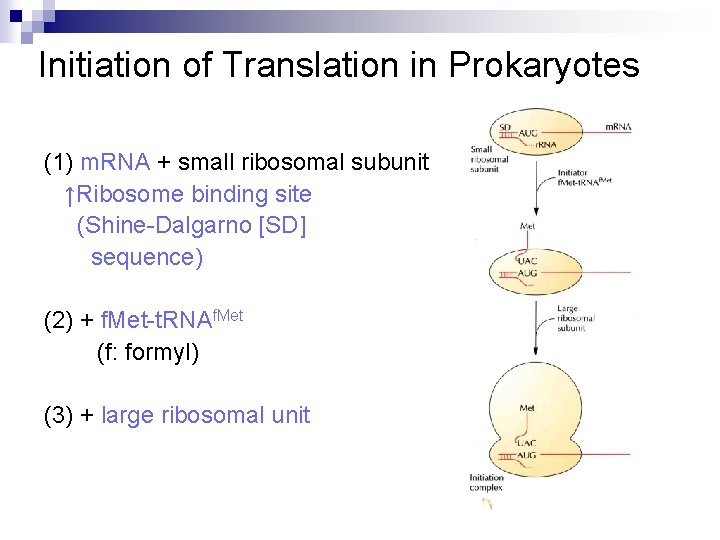
Initiation of Translation in Prokaryotes (1) m. RNA + small ribosomal subunit ↑Ribosome binding site (Shine-Dalgarno [SD] sequence) (2) + f. Met-t. RNAf. Met (f: formyl) (3) + large ribosomal unit
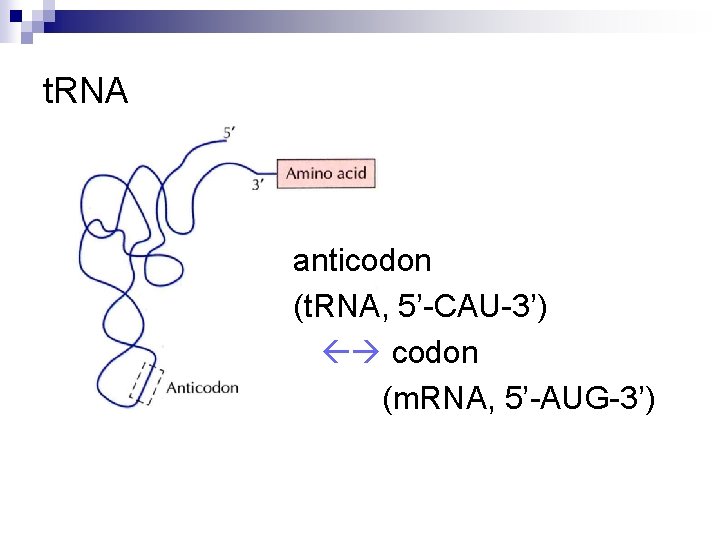
t. RNA anticodon (t. RNA, 5’-CAU-3’) codon (m. RNA, 5’-AUG-3’)
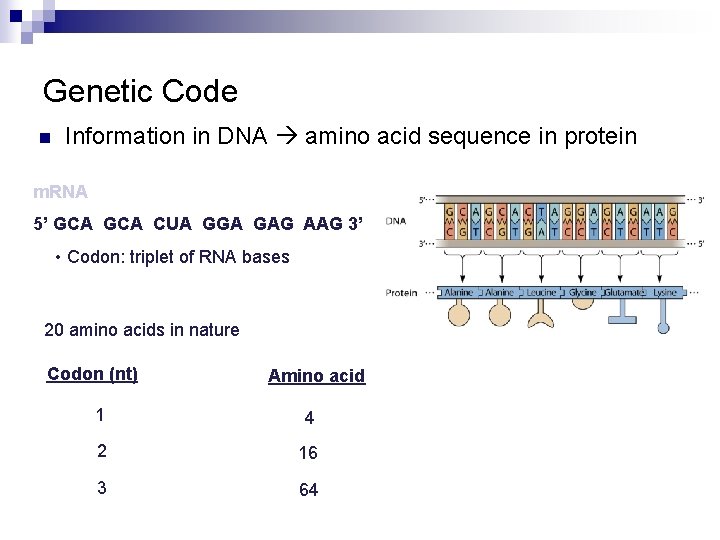
Genetic Code n Information in DNA amino acid sequence in protein m. RNA 5’ GCA CUA GGA GAG AAG 3’ • Codon: triplet of RNA bases 20 amino acids in nature Codon (nt) Amino acid 1 4 2 16 3 64
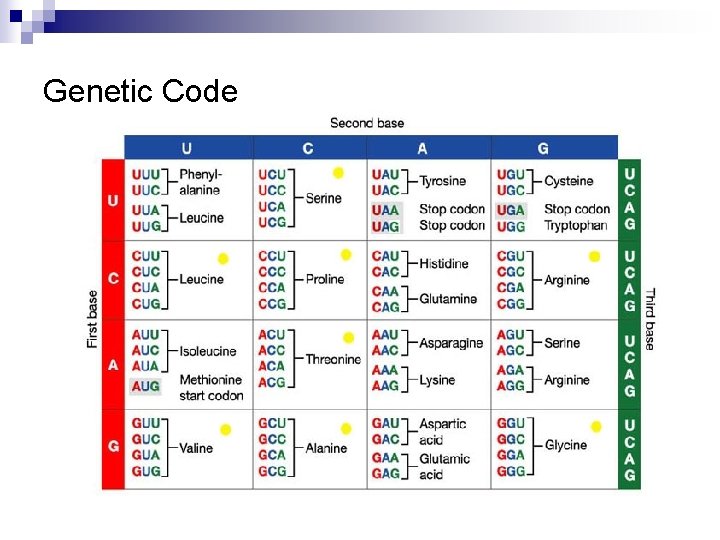
Genetic Code
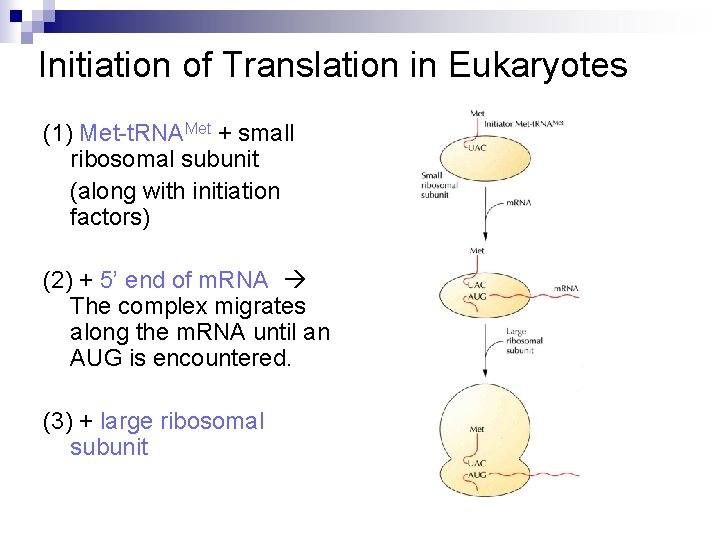
Initiation of Translation in Eukaryotes (1) Met-t. RNAMet + small ribosomal subunit (along with initiation factors) (2) + 5’ end of m. RNA The complex migrates along the m. RNA until an AUG is encountered. (3) + large ribosomal subunit
Translation Initiation n Elongation n Termination n ¨ Elongation and termination phases are very similar in prokaryotes and eukaryotes.
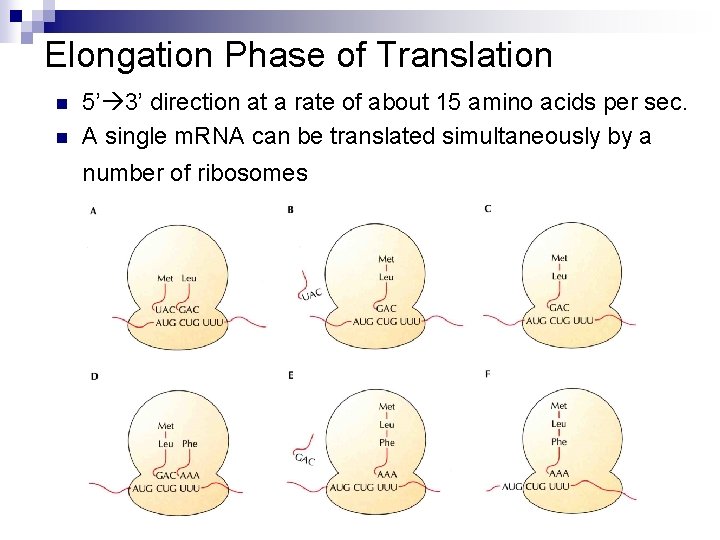
Elongation Phase of Translation n n 5’ 3’ direction at a rate of about 15 amino acids per sec. A single m. RNA can be translated simultaneously by a number of ribosomes

n Table 3. 2 Genetic code and codon usage
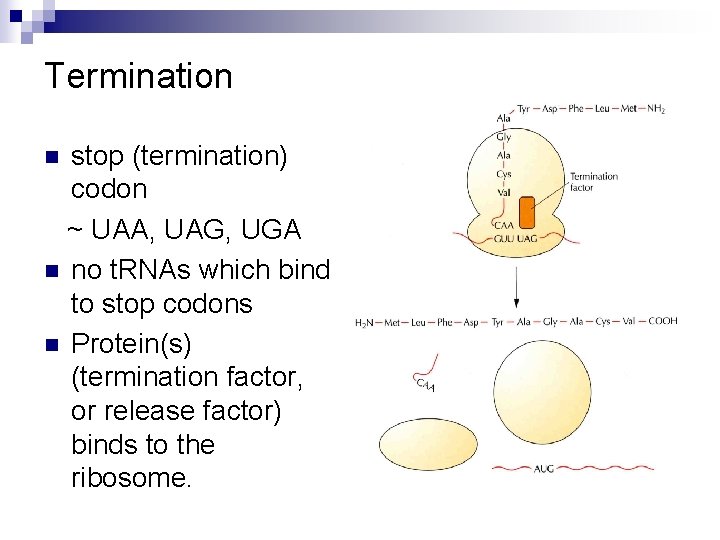
Termination stop (termination) codon ~ UAA, UAG, UGA n no t. RNAs which bind to stop codons n Protein(s) (termination factor, or release factor) binds to the ribosome. n

n In most proteins, the methionine at the Nterminus is cleaved off, leaving the second encoded amino acid as the N-terminal moiety.
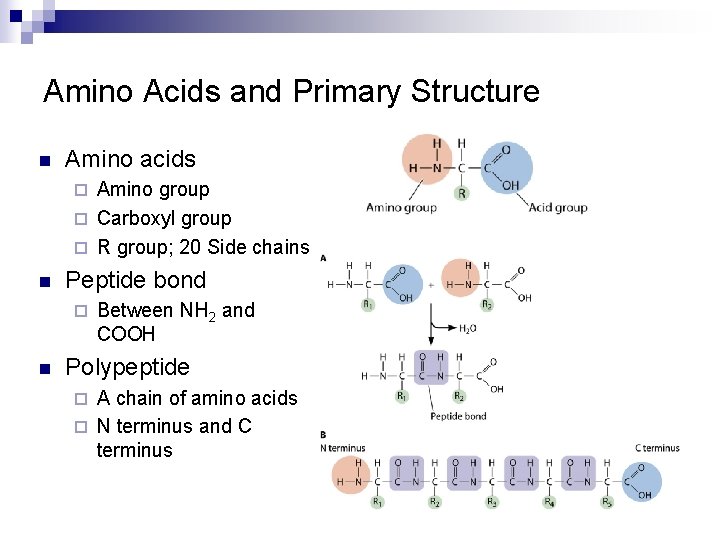
Amino Acids and Primary Structure n Amino acids Amino group ¨ Carboxyl group ¨ R group; 20 Side chains ¨ n Peptide bond ¨ n Between NH 2 and COOH Polypeptide A chain of amino acids ¨ N terminus and C terminus ¨
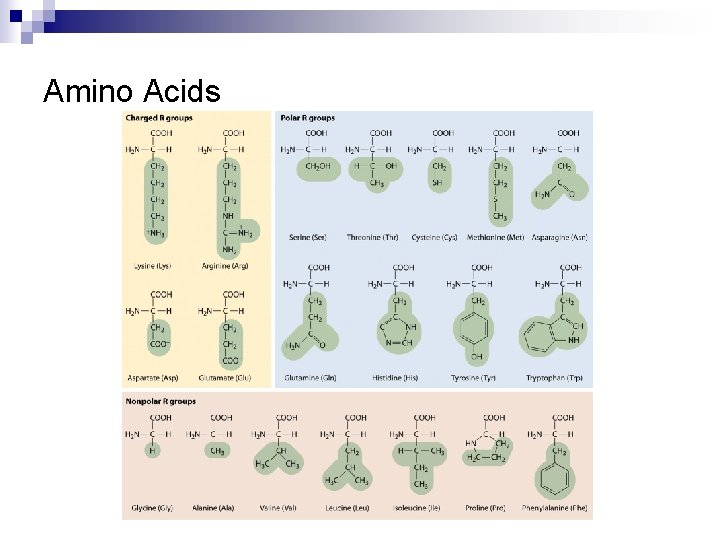
Amino Acids

Regulation of Transcription in Bacteria
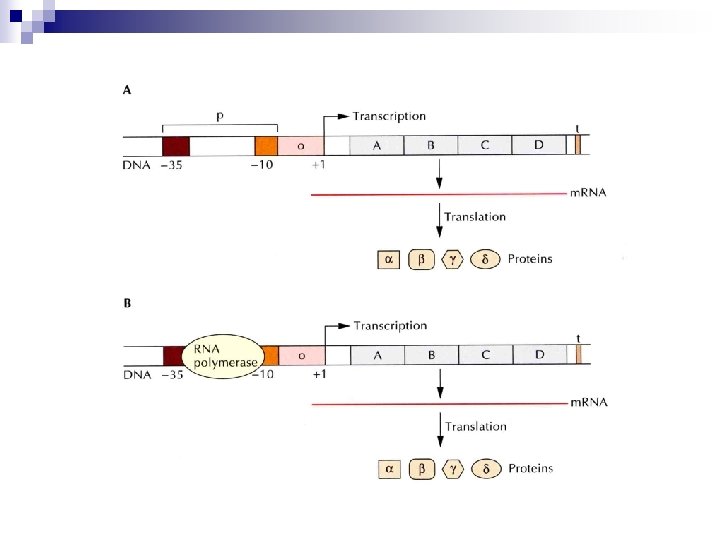

n Operon ¨ Frequently, bacterial structural genes that encode proteins for a single metabolic pathway are contiguous. This arrangement is called an operon. n Promoter region ¨ -10 region (TATAAT, TATA box, Pribnow box) ¨ -35 region (TTGAC sequence)
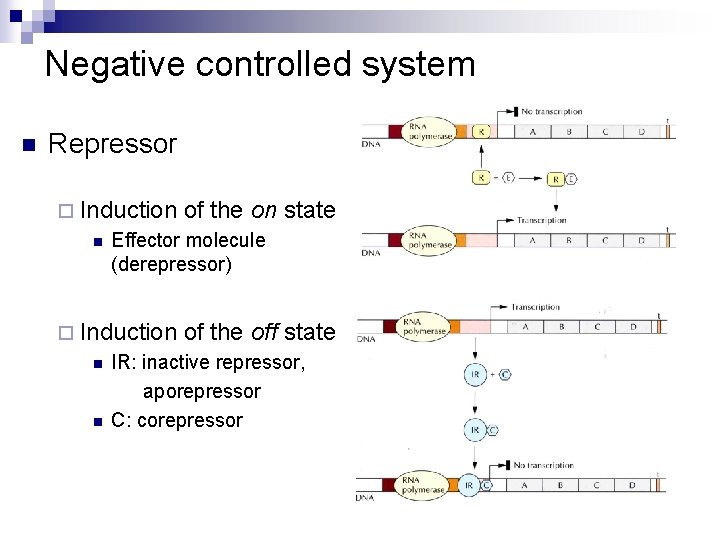
Negative controlled system n Repressor ¨ Induction n Effector molecule (derepressor) ¨ Induction n n of the on state of the off state IR: inactive repressor, aporepressor C: corepressor
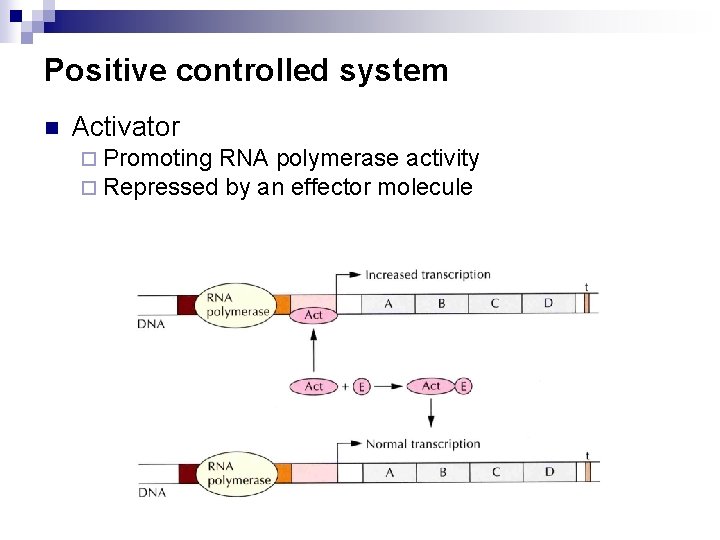
Positive controlled system n Activator ¨ Promoting RNA polymerase activity ¨ Repressed by an effector molecule

Regulation of Transcription in Eukaryotes
n Unlike the situation in prokaryotes, operons are almost never found in the genomes of eukaryotes. In addition to DNA-protein interactions, protein-protein associations are important for regulating eukaryotic transcription. n Promoter region ¨ -25 n bp (TATA box, or Hogness box) 8 nucleotides including TATA sequence ¨ -75 bp (CCAAT sequence, “cat” box) ¨ -90 bp (GC box --- repeated GC)
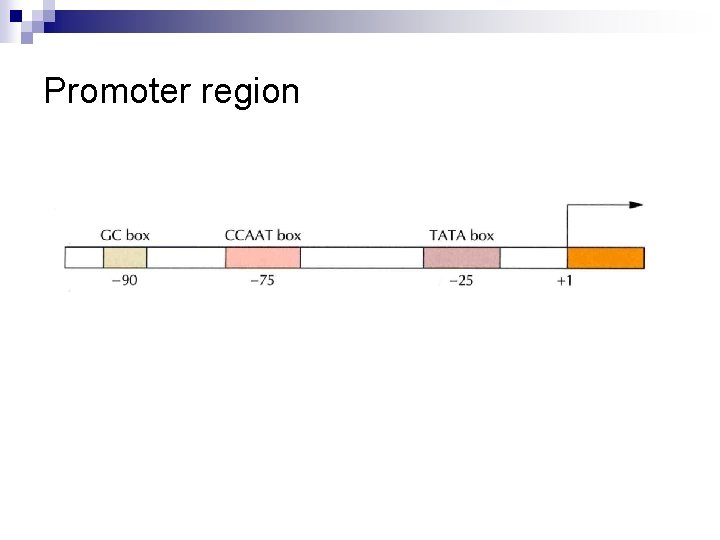
Promoter region
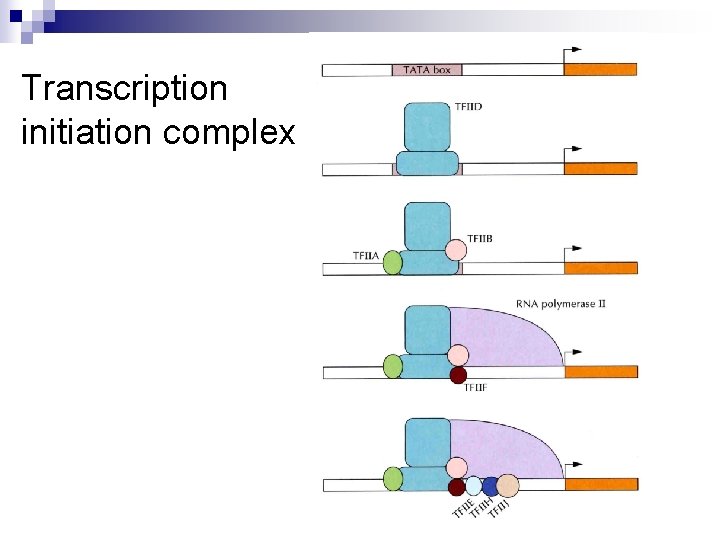
Transcription initiation complex
- Slides: 31