TRANSCRIPTION RNA CLASSIFICATION ribosomal RNA r RNA messenger

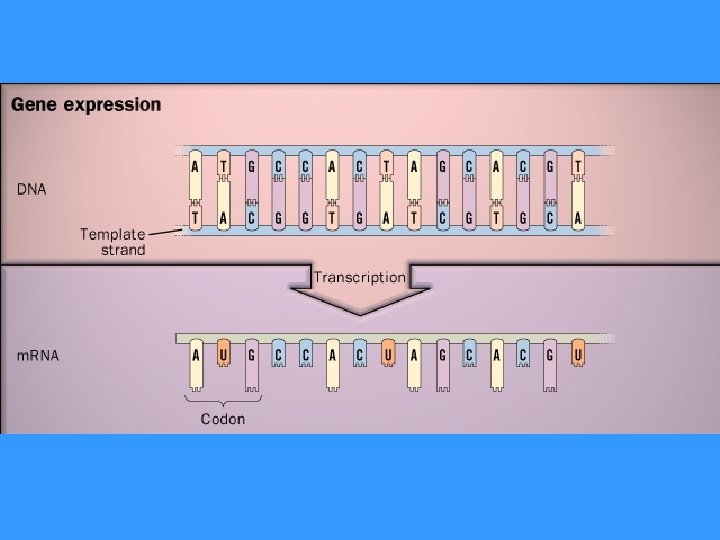
TRANSCRIPTION


RNA CLASSIFICATION • • ribosomal RNA (r. RNA) messenger RNA (m. RNA) transfer RNA (t. RNA) Small nuclear RNA i micro RNA (sn RNA and mi. RNA) • r, m and t RNA included in protein synthesis • sn and mi RNA involved in splicing of m. RNA and modulation of gene expression changing the function of m. RNA


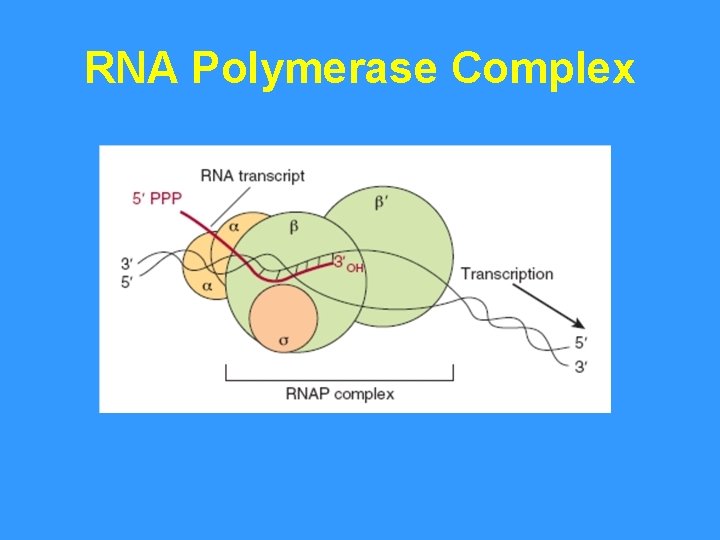
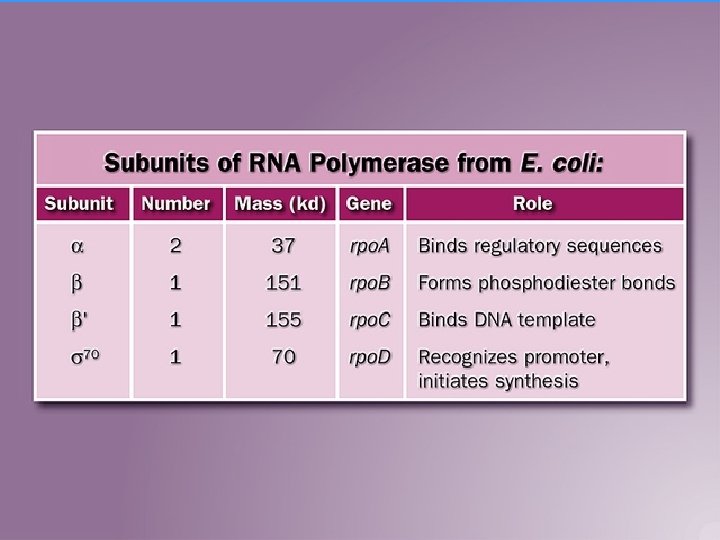
RNA Polymerase Complex
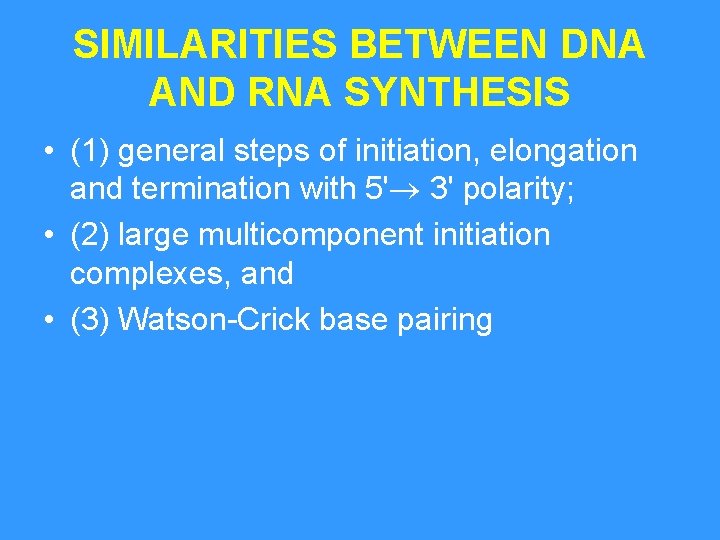
SIMILARITIES BETWEEN DNA AND RNA SYNTHESIS • (1) general steps of initiation, elongation and termination with 5' 3' polarity; • (2) large multicomponent initiation complexes, and • (3) Watson-Crick base pairing

DIFFERENCES BETWEEN DNA AND RNA SYNTHESIS (1) RNA- ribonucleotides instead deoxyribonucleotides; (2) U instead T pairing with A in RNA; (3) Primers are not involved in RNA synthesis since RNA polymerases have ability of initiating de novo synthesis; (4) Only part of RNA genome is transcribed, while whole genome is completely copied during DNA replication, and (5) No proofreading during RNA transcription
RNA IS SYNTHESIZED FROM A DNA TEMPLATE BY AN RNA POLYMERASE
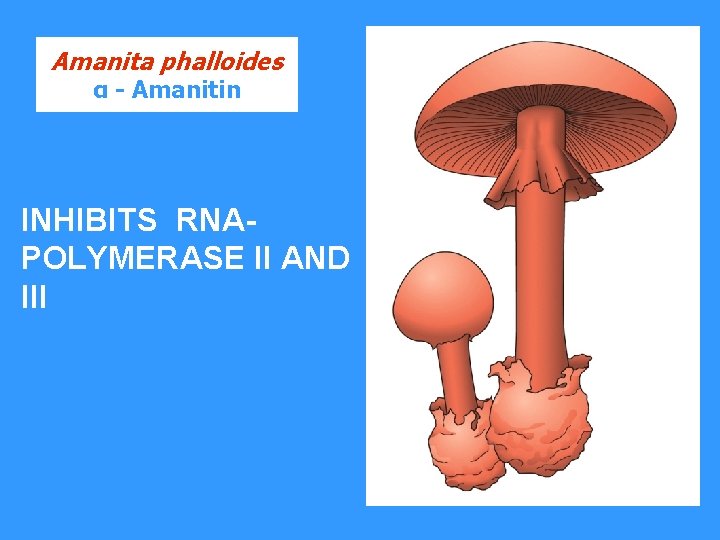
Amanita phalloides α - Amanitin INHIBITS RNAPOLYMERASE II AND III
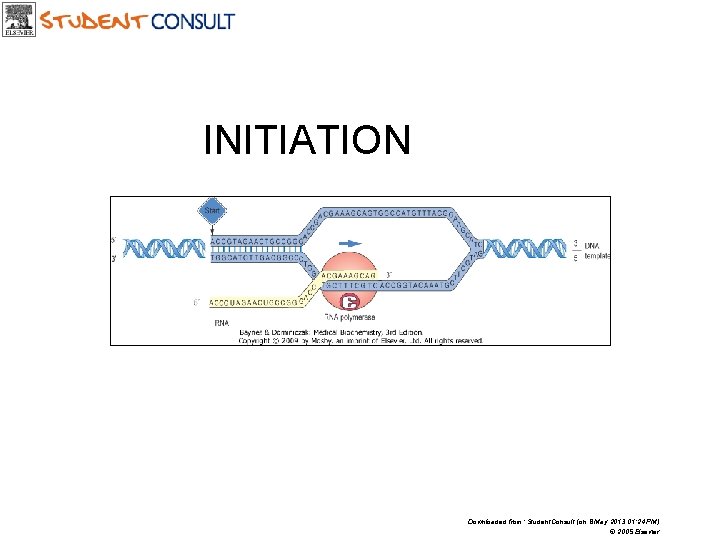
INITIATION Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
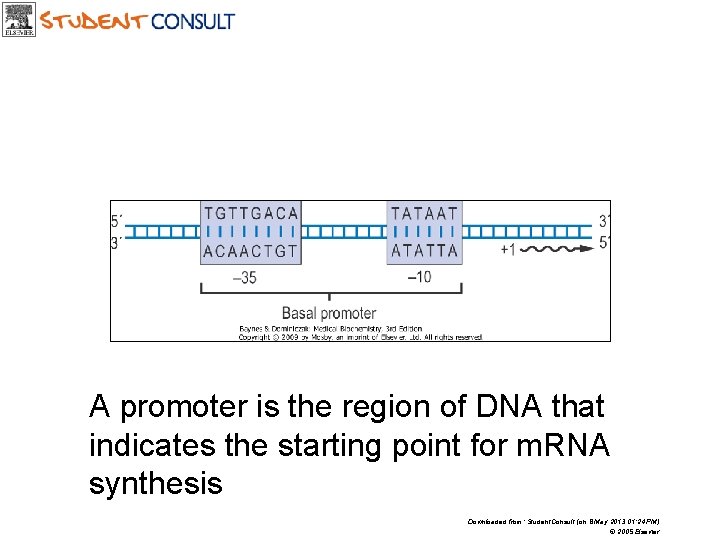
A promoter is the region of DNA that indicates the starting point for m. RNA synthesis Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
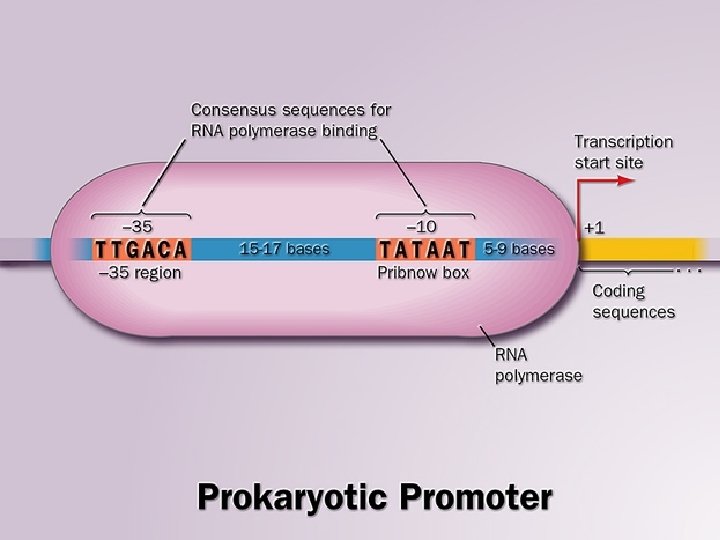
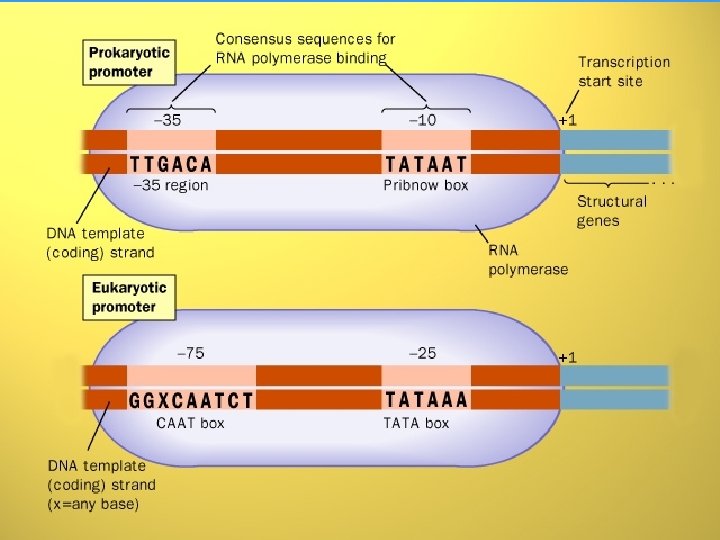
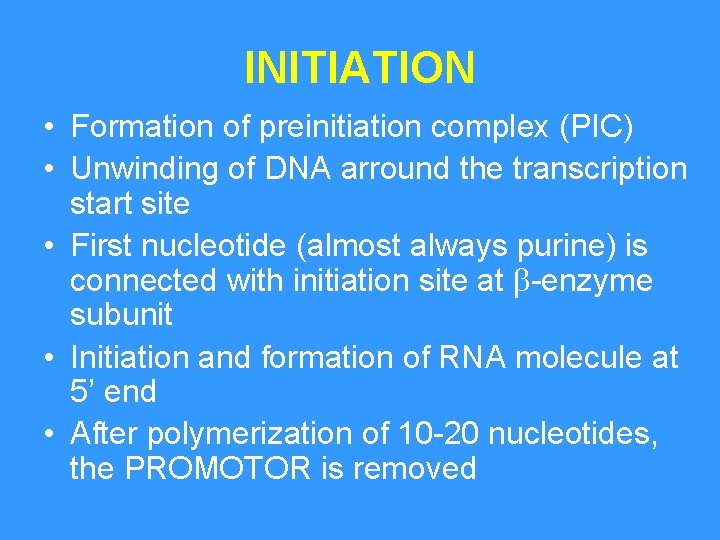
INITIATION • Formation of preinitiation complex (PIC) • Unwinding of DNA arround the transcription start site • First nucleotide (almost always purine) is connected with initiation site at -enzyme subunit • Initiation and formation of RNA molecule at 5’ end • After polymerization of 10 -20 nucleotides, the PROMOTOR is removed
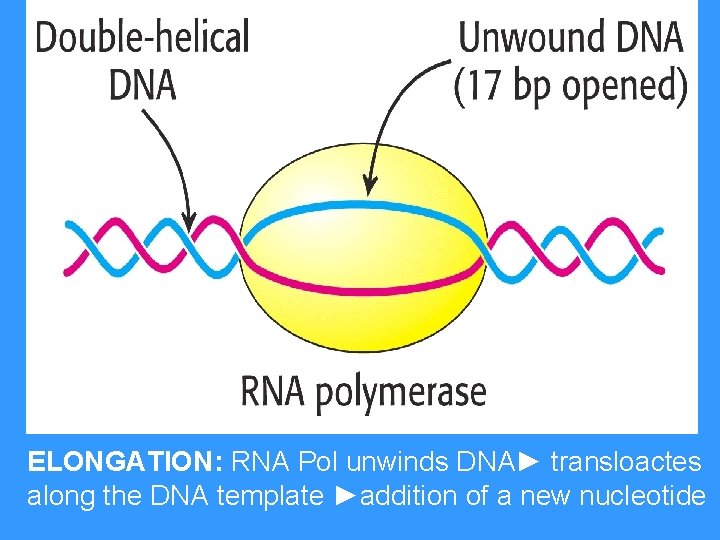
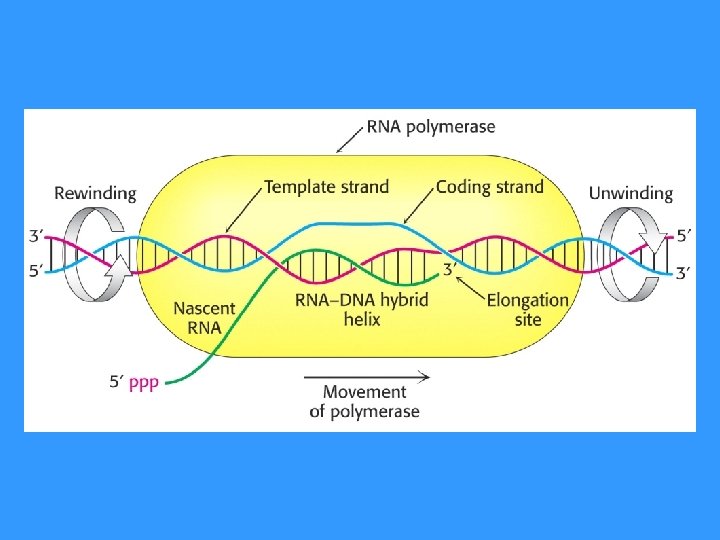
ELONGATION: RNA Pol unwinds DNA► transloactes along the DNA template ►addition of a new nucleotide
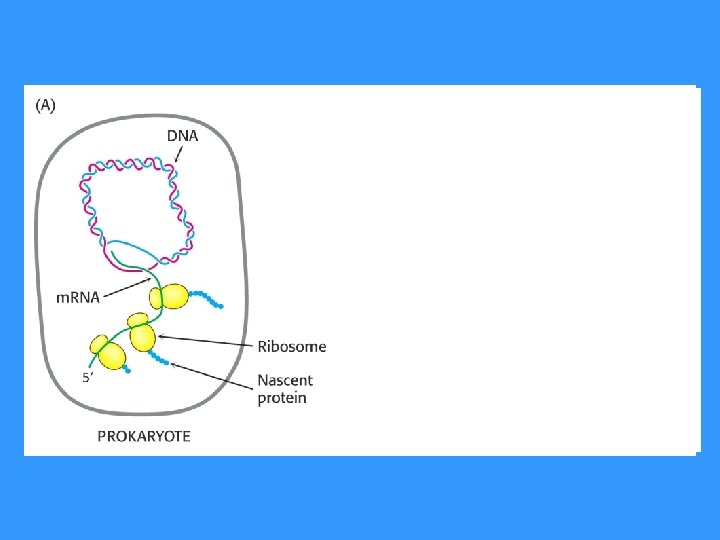
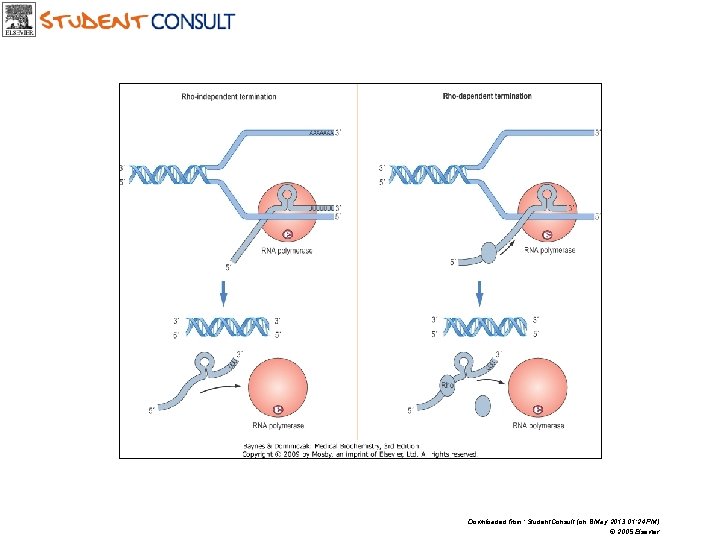
TERMINATION IN PROKARYOTES • Termination of RNA synthesis in prokaryotes is signalized by the sequence in DNA template strand by signal which recognize termination protein – rho factor ( ).

TERMINATION • Rho- independent termination: a hairpin loop formed in the RNA upstream of 6 -8 uridine residues near the 3’ end ►RNA Pol dislodged • Rho- dependent termination: when RNA Pol encounters the hairpin loop, the rho protein (an ATP dependent helicase) moves along the RNA, catches up with RNA Pol and dislodges it.
Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
TERMINATION IN EUKARYOTES • Termination of RNA synthesis in eukaryotes : RNA Pol I transcript termination factor 1 (TTF-1) binds to the terminator site (-1000) ► when RNA Pol I meets TTF-1, a releasing factor catalyzes RNA Pol release
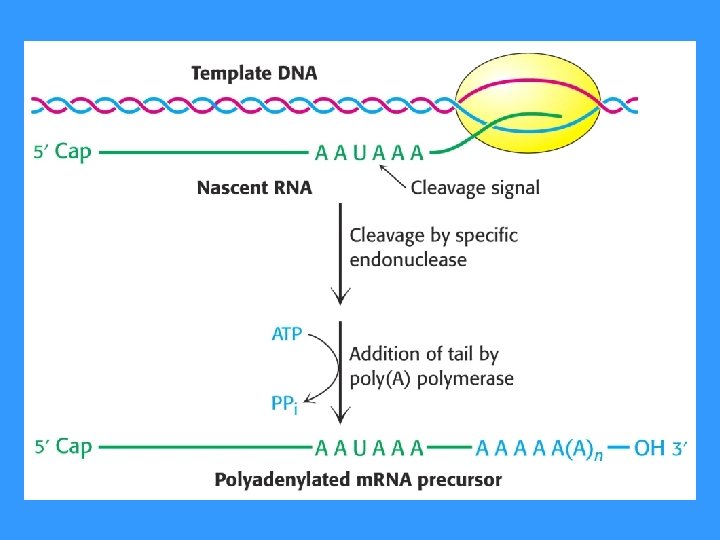
RNA PROCESSING • RNA MOLECULES ARE USUALLY PROCESSED BEFORE THEY BECOME FUNCTIONAL • • • CAP (5’) Poly A (3’) SPLICING
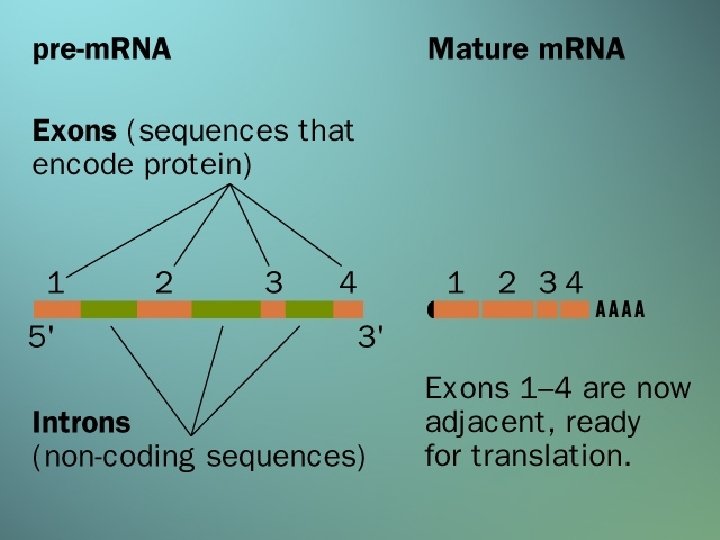
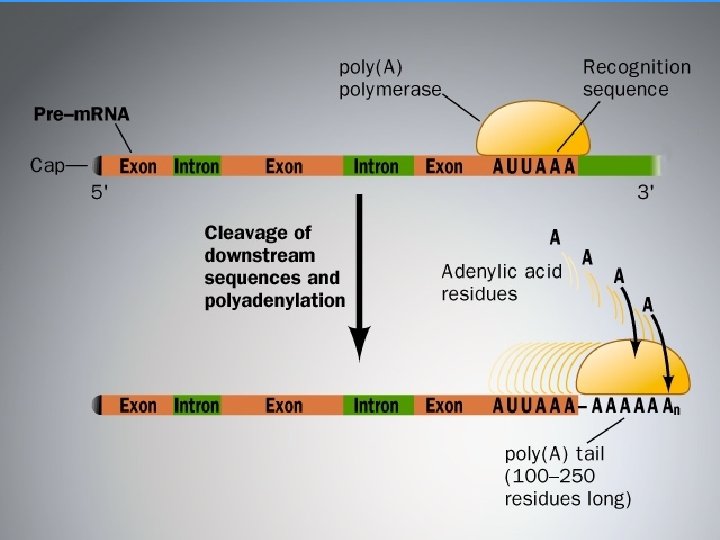
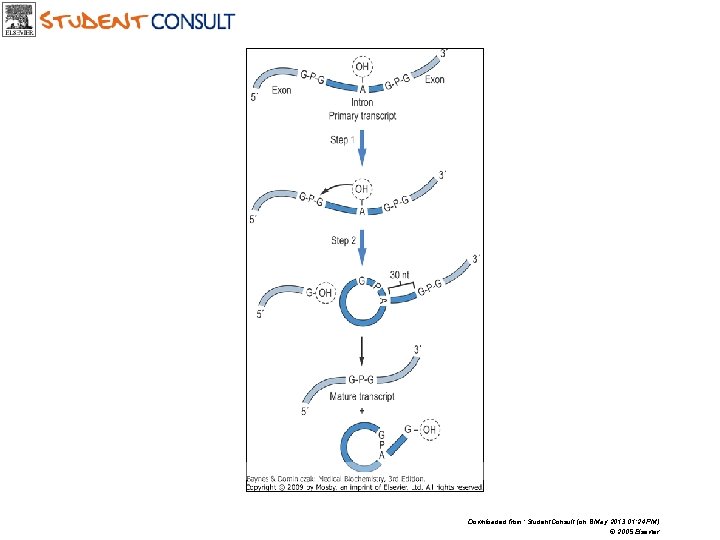
Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier

ALTERNATIVE SPLICING enables formation of a different m. RNA (different proteins are functional in a different tissues)
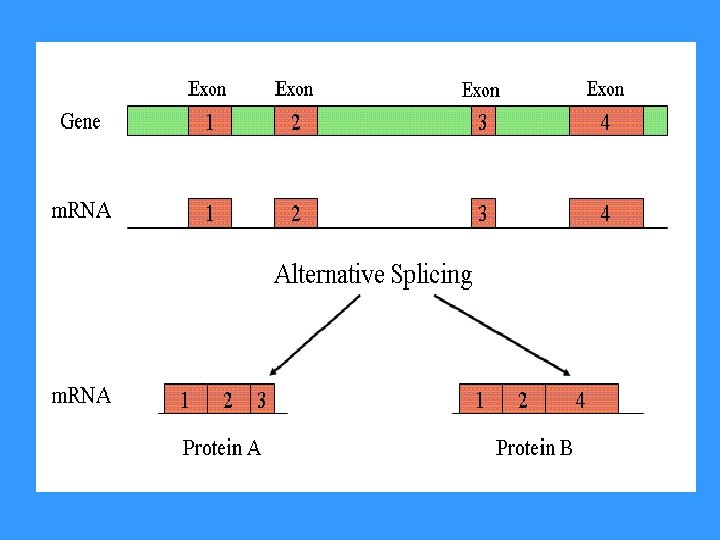
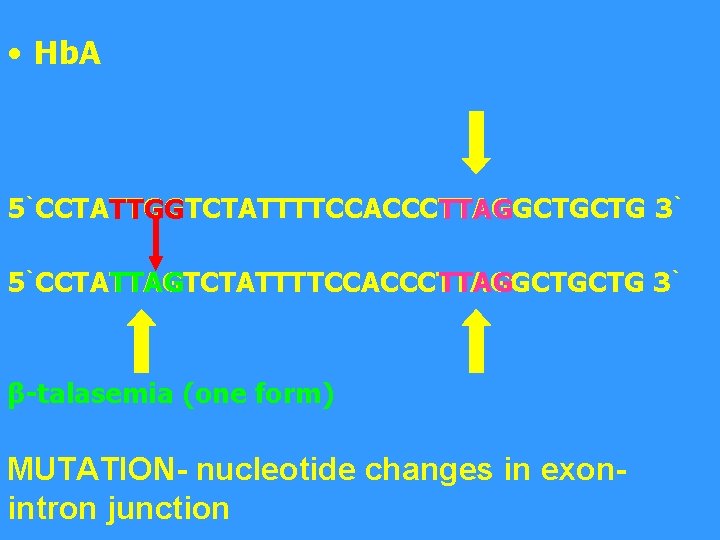
• Hb. A 5`CCTATTGGTCTATTTTCCACCCTTAGGCTGCTG 3` TTGG TTAG 5`CCTATTAGTCTATTTTCCACCCTTAGGCTGCTG 3` TTAG β-talasemia (one form) MUTATION- nucleotide changes in exonintron junction
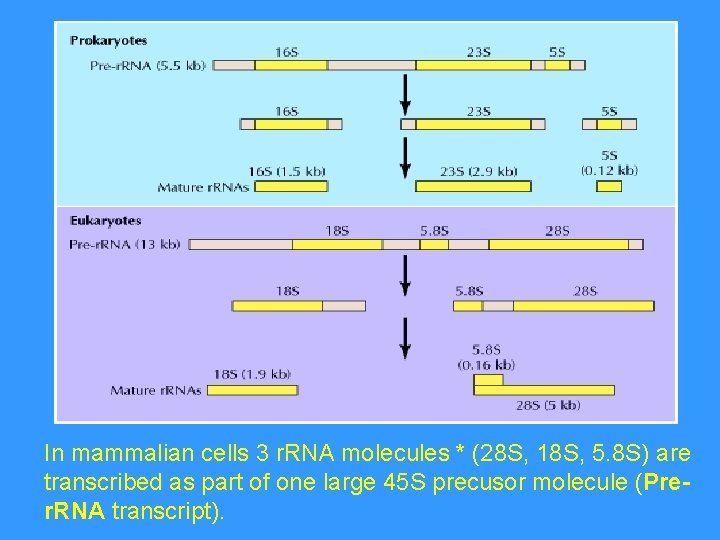
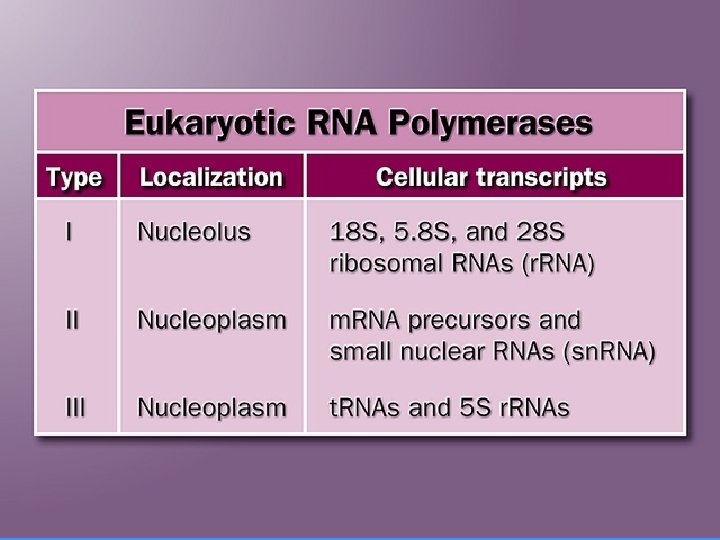
In mammalian cells 3 r. RNA molecules * (28 S, 18 S, 5. 8 S) are transcribed as part of one large 45 S precusor molecule (Prer. RNA transcript).
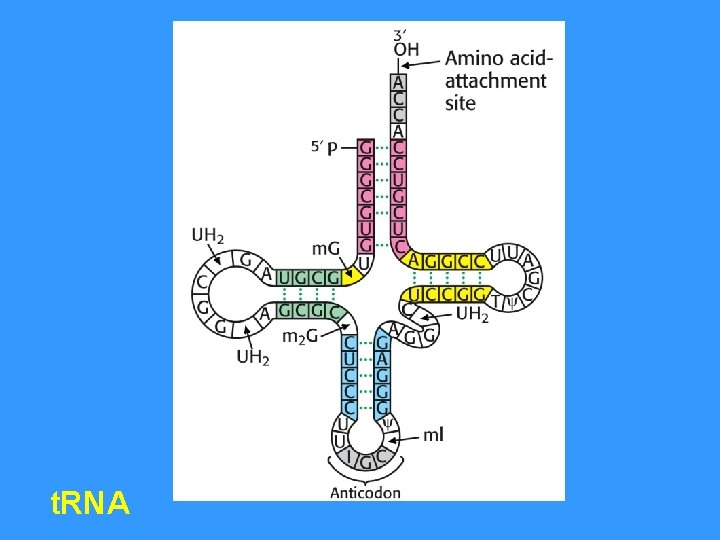
t. RNA
Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
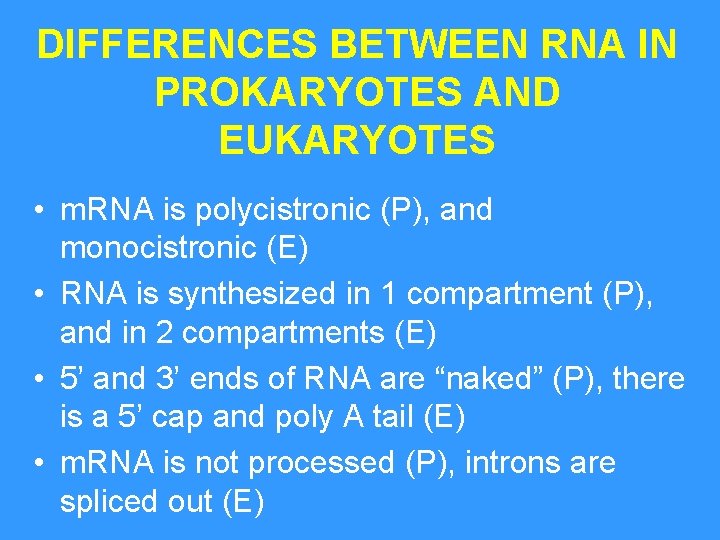
DIFFERENCES BETWEEN RNA IN PROKARYOTES AND EUKARYOTES • m. RNA is polycistronic (P), and monocistronic (E) • RNA is synthesized in 1 compartment (P), and in 2 compartments (E) • 5’ and 3’ ends of RNA are “naked” (P), there is a 5’ cap and poly A tail (E) • m. RNA is not processed (P), introns are spliced out (E)

TRANSLATION
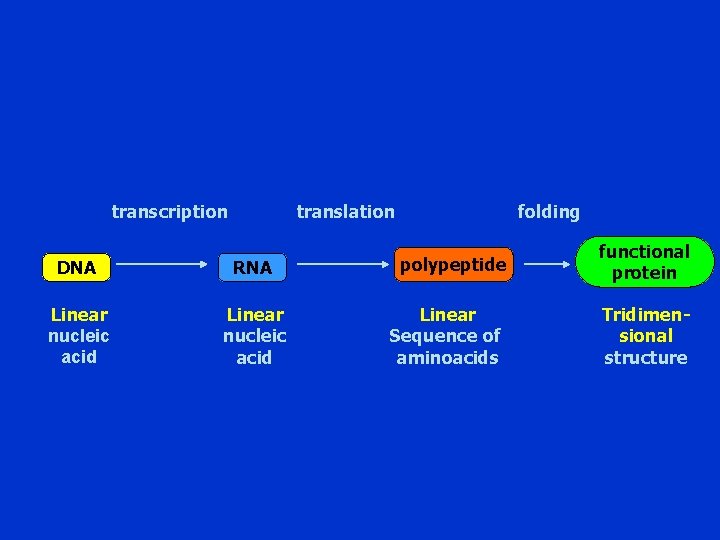
transcription translation DNA RNA Linear nucleic acid folding polypeptide Linear Sequence of aminoacids functional protein Tridimensional structure
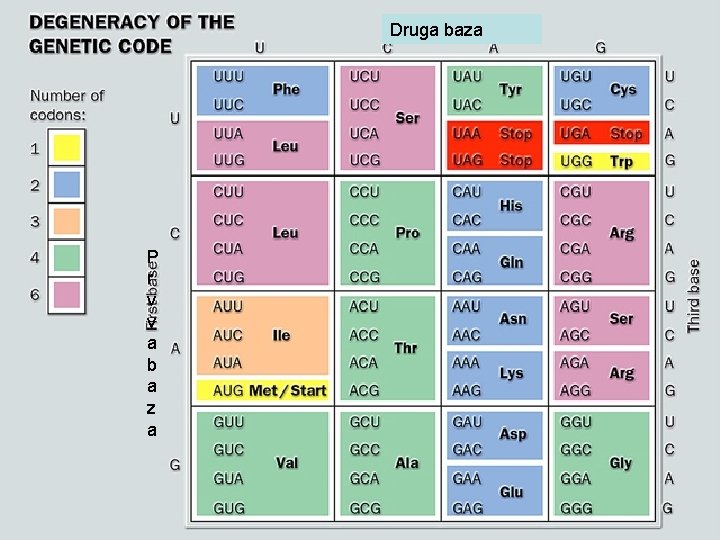
Druga baza P r v v a b a z a
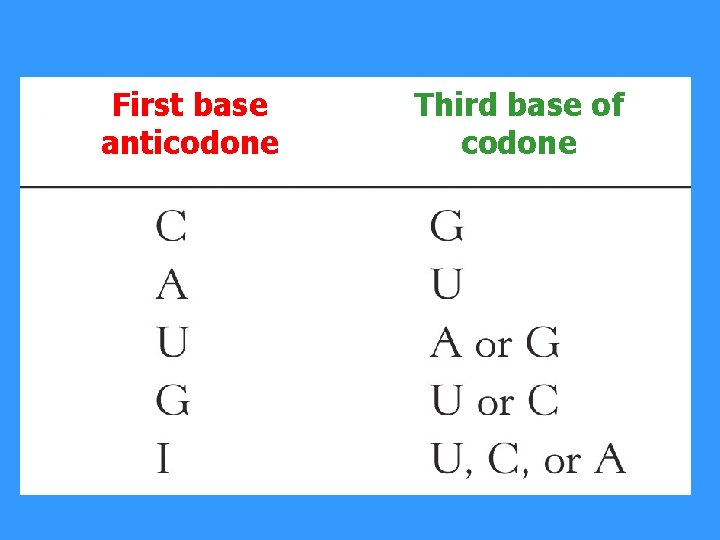
First base anticodone Third base of codone
Like transcription, protein synthesis could be described with three phases : • INITIATION , • ELONGATION AND • TERMINATION
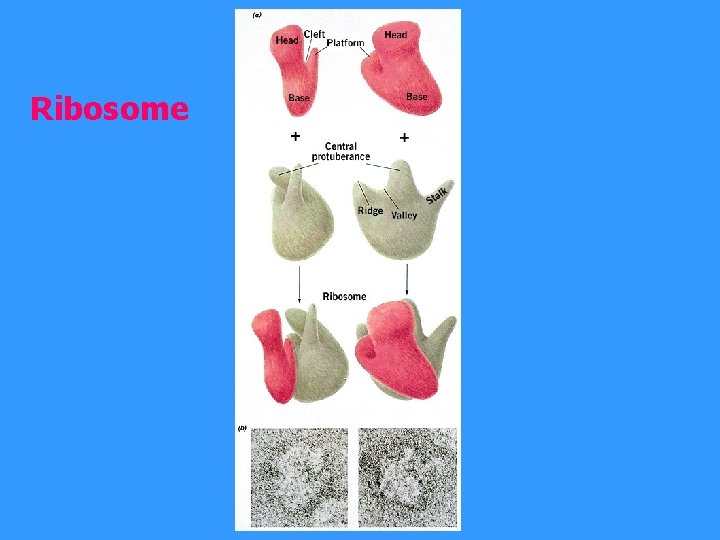
Ribosome
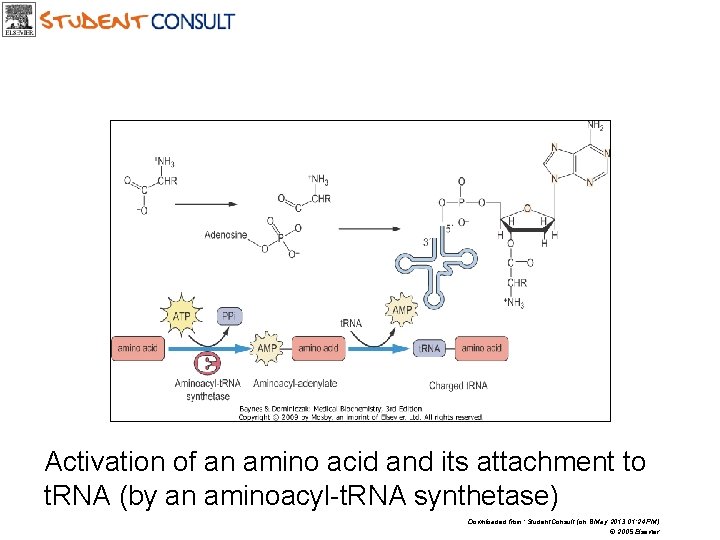
Activation of an amino acid and its attachment to t. RNA (by an aminoacyl-t. RNA synthetase) Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
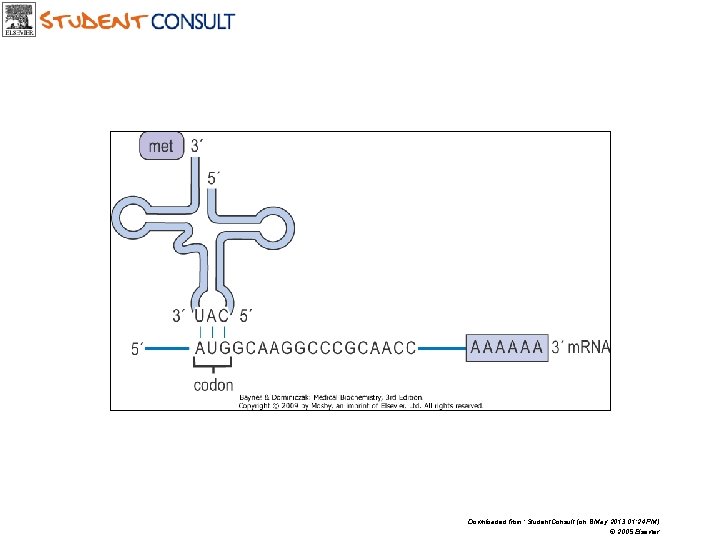
Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
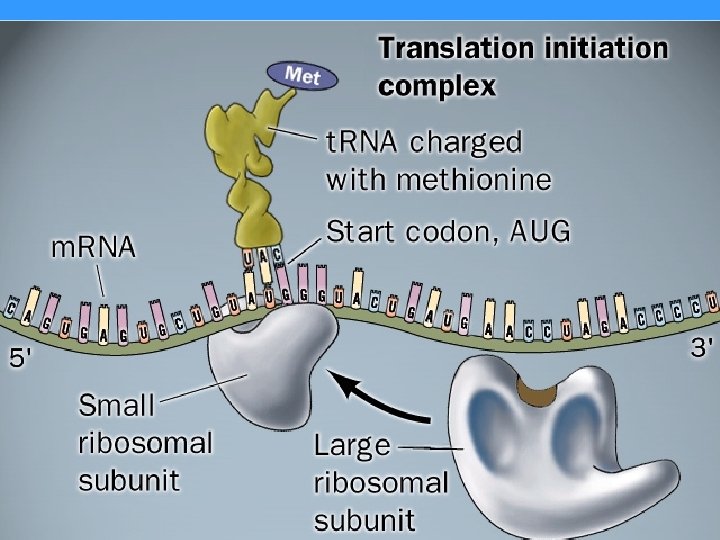
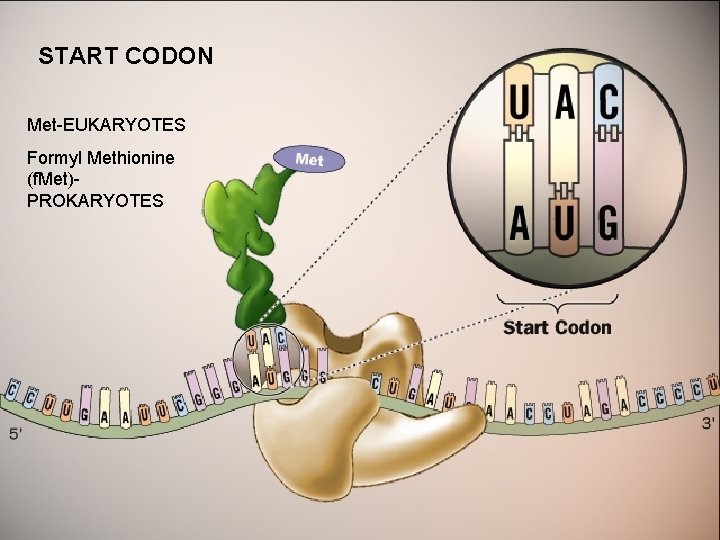
START CODON Met-EUKARYOTES Formyl Methionine (f. Met)PROKARYOTES
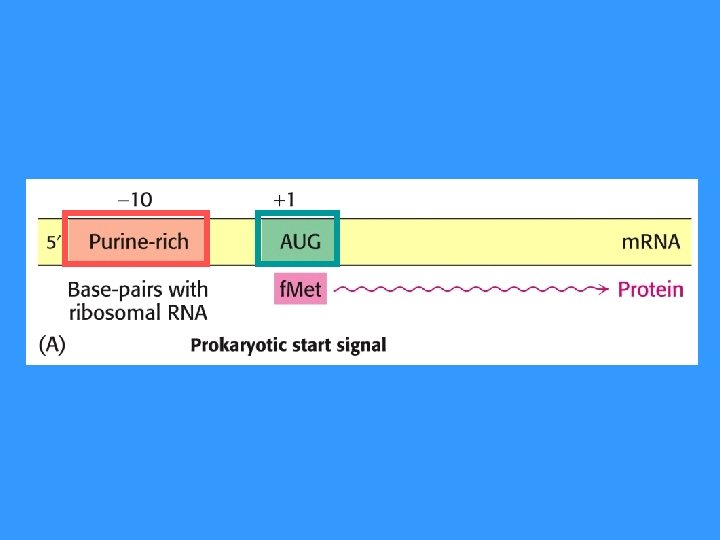
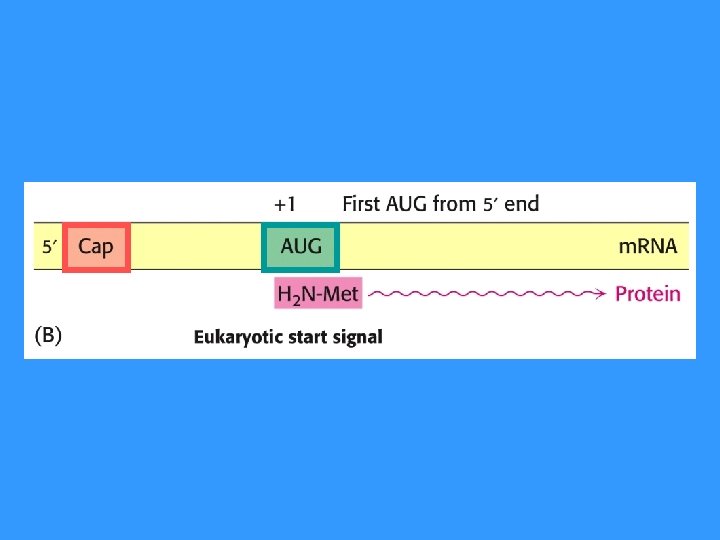
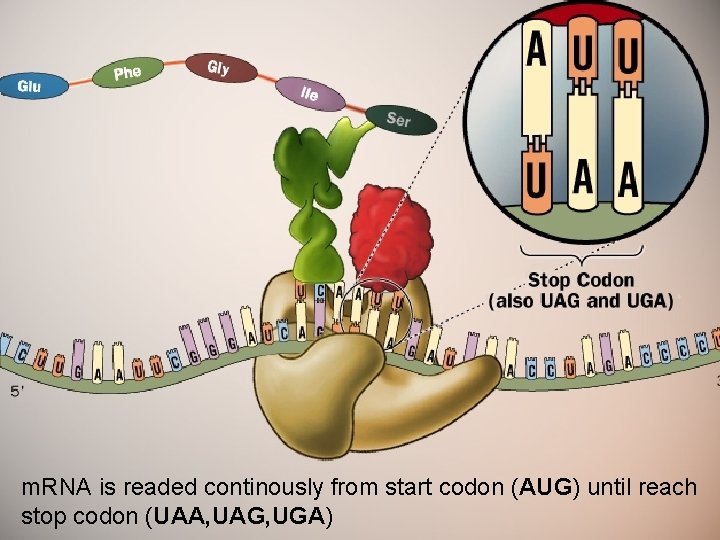
m. RNA is readed continously from start codon (AUG) until reach stop codon (UAA, UAG, UGA)
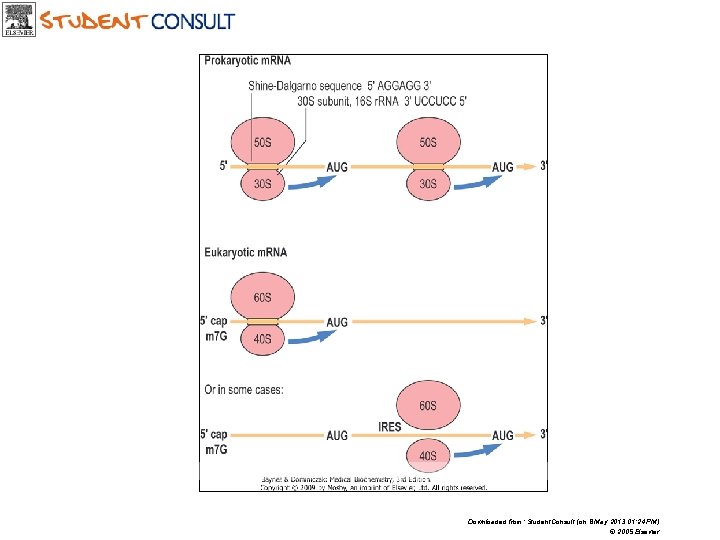
Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
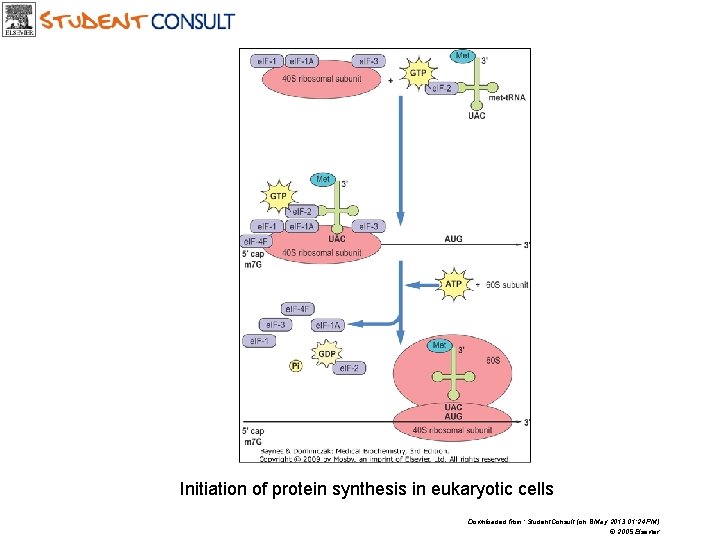
Initiation of protein synthesis in eukaryotic cells Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
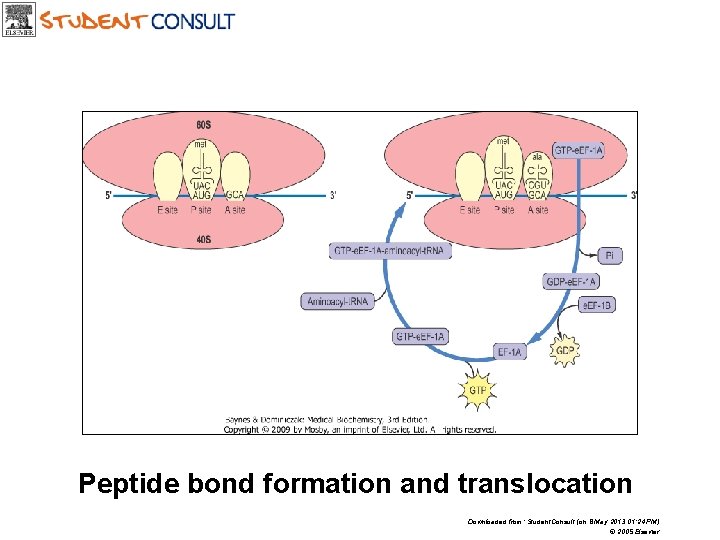
Peptide bond formation and translocation Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
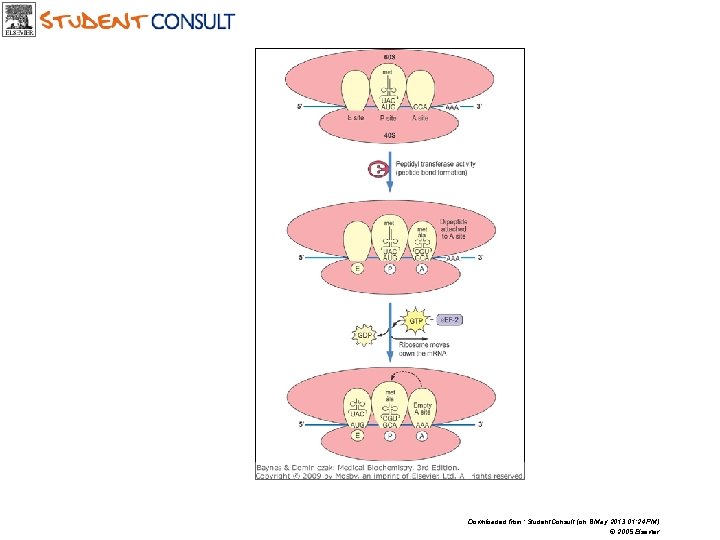
Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
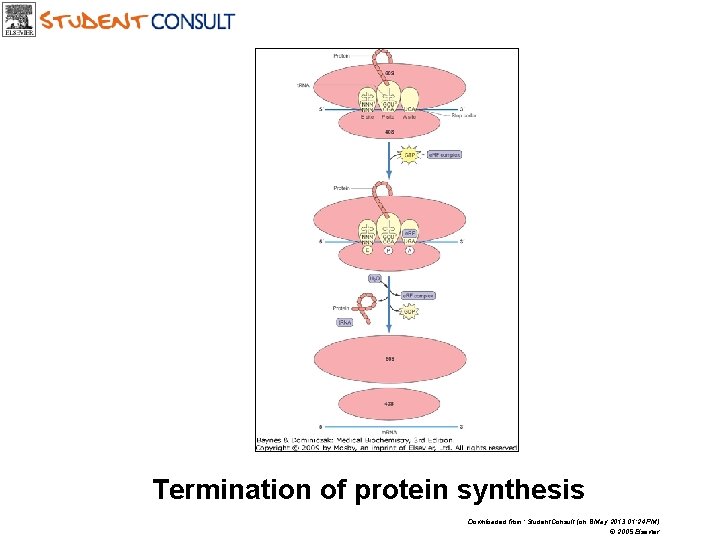
Termination of protein synthesis Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
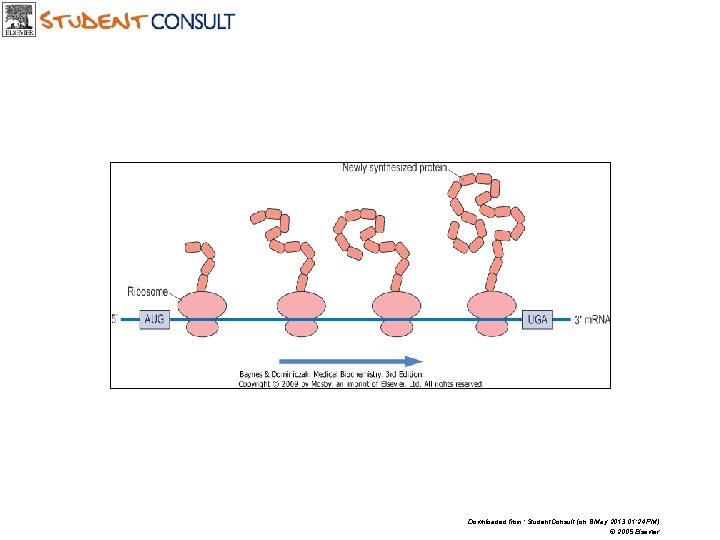
Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier
CELLULAR PROTEIN DEGRADATION • Proteasome destroys intracellular defective, misfolded proteins • Proteasome is an ATP- dependent protease complex present in the cytosol and in the nucleus • Ubiqutin cycle is involved in marking proteins for degradation by the proteasome (more highly ubiqutinated the protein, the more susceptible it is to proteasomal degradation
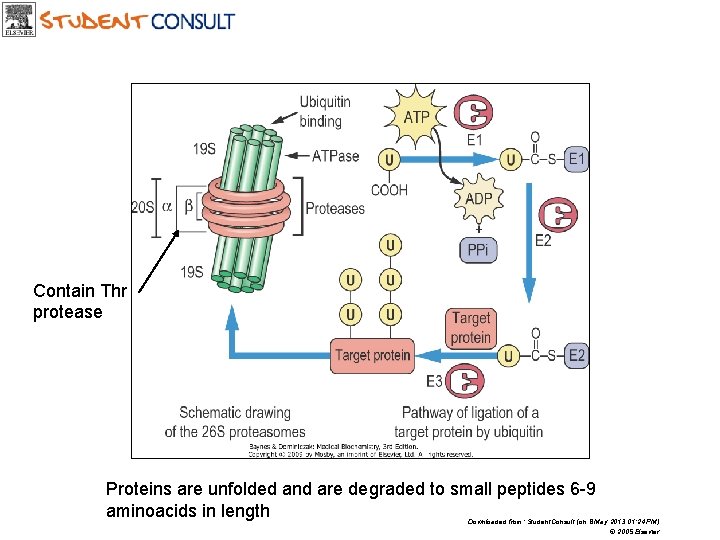
Contain Thr protease Proteins are unfolded and are degraded to small peptides 6 -9 aminoacids in length Downloaded from: Student. Consult (on 8 May 2013 01: 24 PM) © 2005 Elsevier

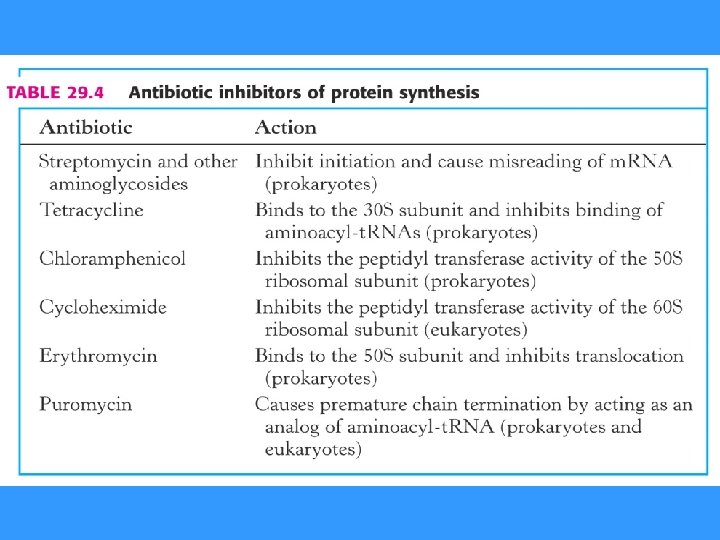
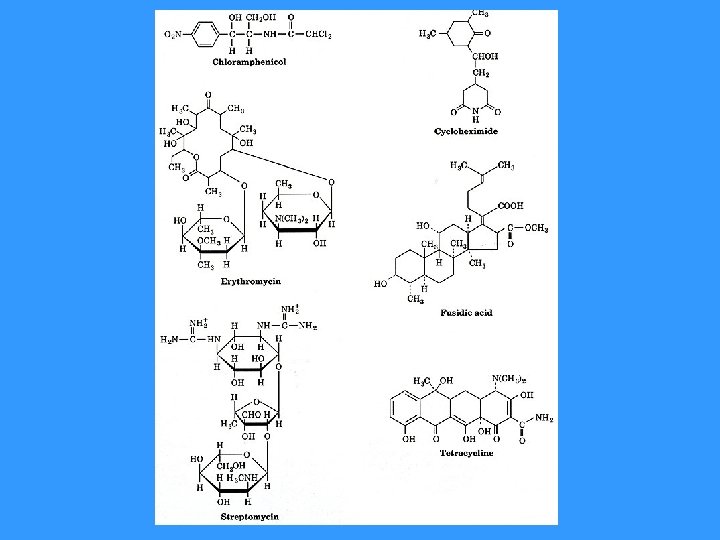
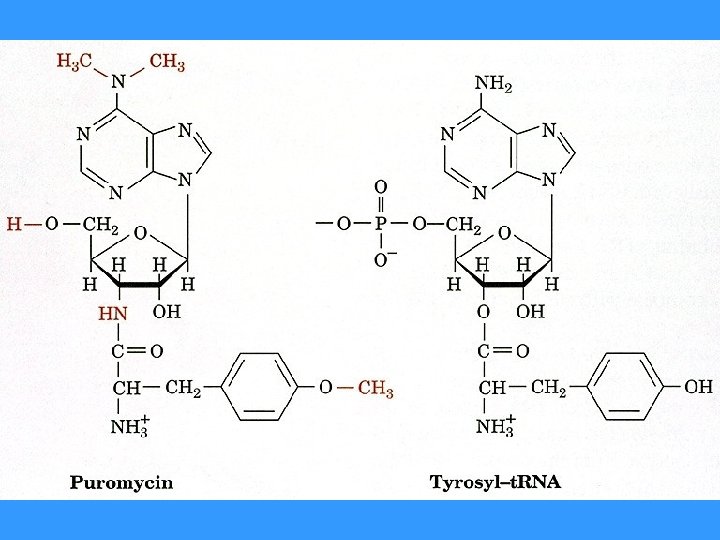
INHIBITORS OF PROTEIN SYNTHESIS
- Slides: 62