Transcription and Posttranscriptional Modifications in Prokaryotes and Eukaryotes









- Slides: 69

Transcription and Posttranscriptional Modifications in Prokaryotes and Eukaryotes By Amr S Moustafa, MD, Ph. D Medical Biochemistry and Molecular Biology Department Faculty of Medicine, Ain Shams University amrsm@hotmail. com

Intended Learning Outcomes 1. Describe the transcription process in prokaryotes 2. Identify the different phases (stages) of transcription-“Initiation”, “Elongation” and “Termination” 3. Define the promoter and its role in transcription 4. Compare eukaryotic to prokaryotic transcription 5. Identify post-transcriptional modifications of the newly synthesized primary transcript

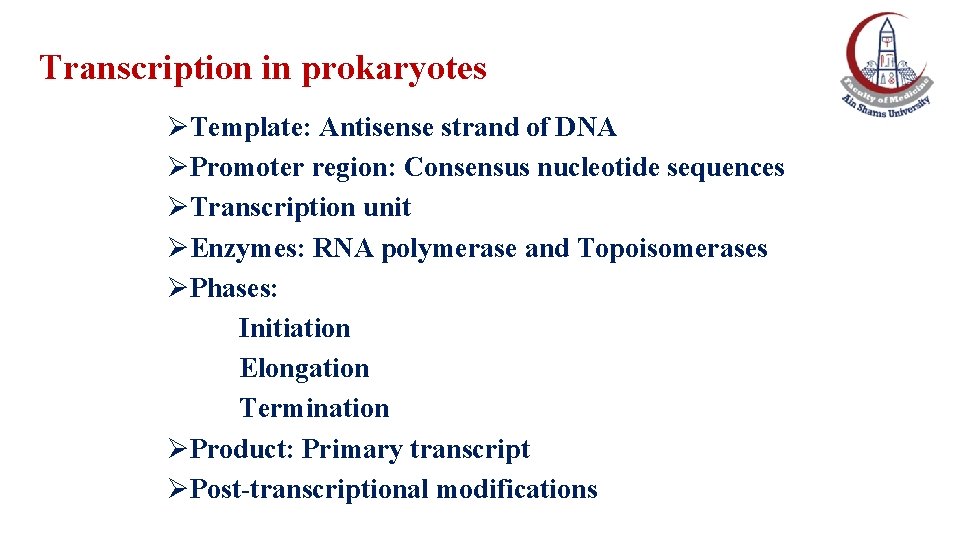
Transcription • It is the synthesis of RNA under the direction of DNA (DNA-directed RNA synthesis), By a DNA-dependent RNA polymerase.

Transcription in prokaryotes ØTemplate: Antisense strand of DNA ØPromoter region: Consensus nucleotide sequences ØTranscription unit ØEnzymes: RNA polymerase and Topoisomerases ØPhases: Initiation Elongation Termination ØProduct: Primary transcript ØPost-transcriptional modifications

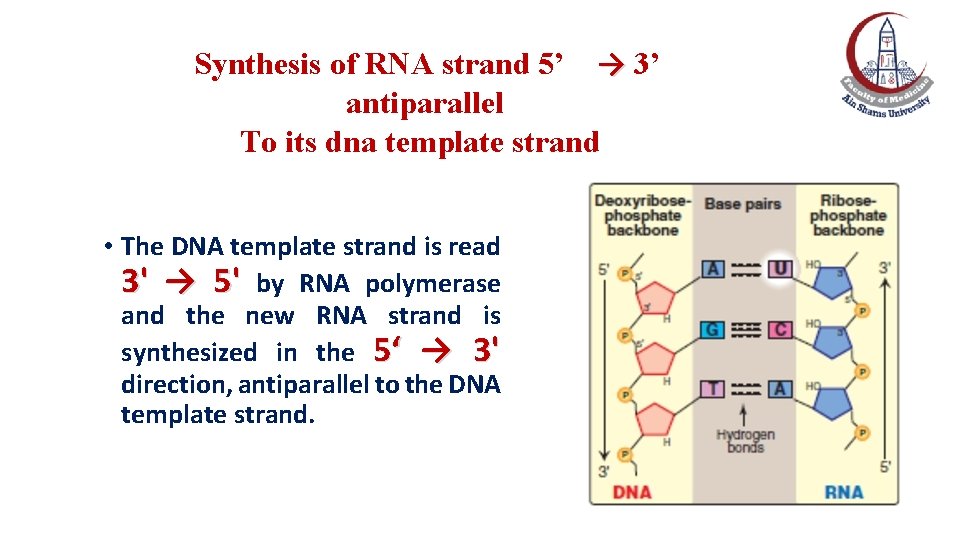
Synthesis of RNA strand 5’ → 3’ antiparallel To its dna template strand • The DNA template strand is read 3' → 5' by RNA polymerase and the new RNA strand is synthesized in the 5‘ → 3' direction, antiparallel to the DNA template strand.

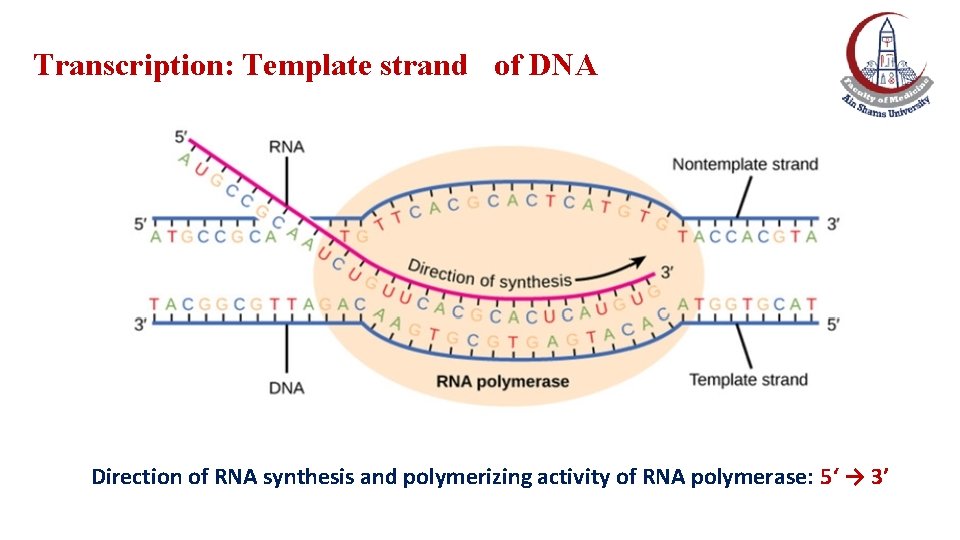
Transcription: Template strand of DNA Direction of RNA synthesis and polymerizing activity of RNA polymerase: 5‘ → 3’

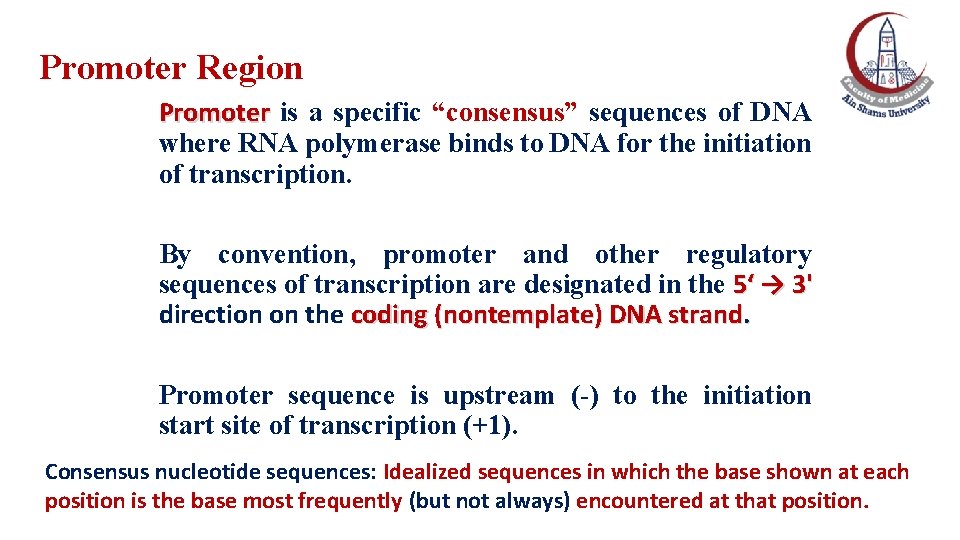
Promoter Region Promoter is a specific “consensus” sequences of DNA where RNA polymerase binds to DNA for the initiation of transcription. By convention, promoter and other regulatory sequences of transcription are designated in the 5‘ → 3' direction on the coding (nontemplate) DNA strand. Promoter sequence is upstream (-) to the initiation start site of transcription (+1). Consensus nucleotide sequences: Idealized sequences in which the base shown at each position is the base most frequently (but not always) encountered at that position.

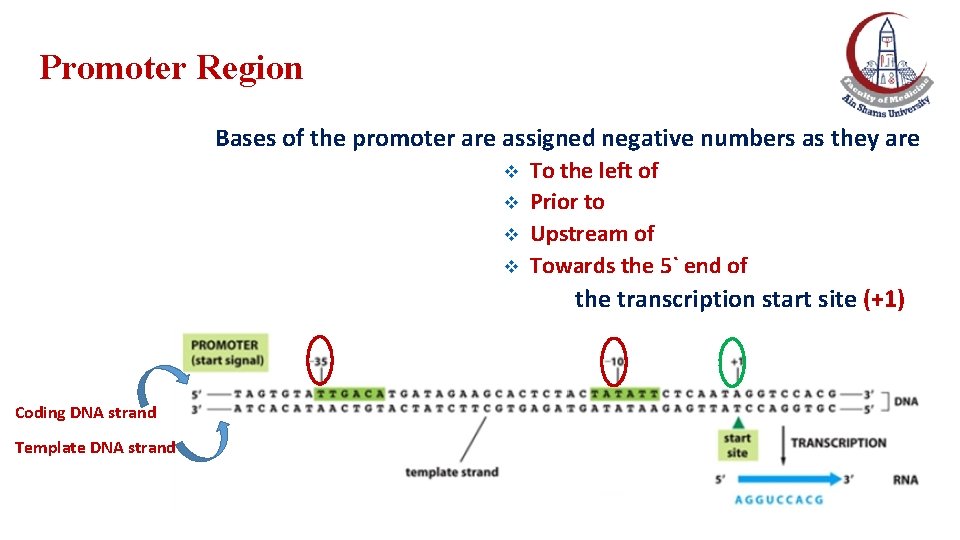
Promoter Region Bases of the promoter are assigned negative numbers as they are v v To the left of Prior to Upstream of Towards the 5` end of the transcription start site (+1) Coding DNA strand Template DNA strand

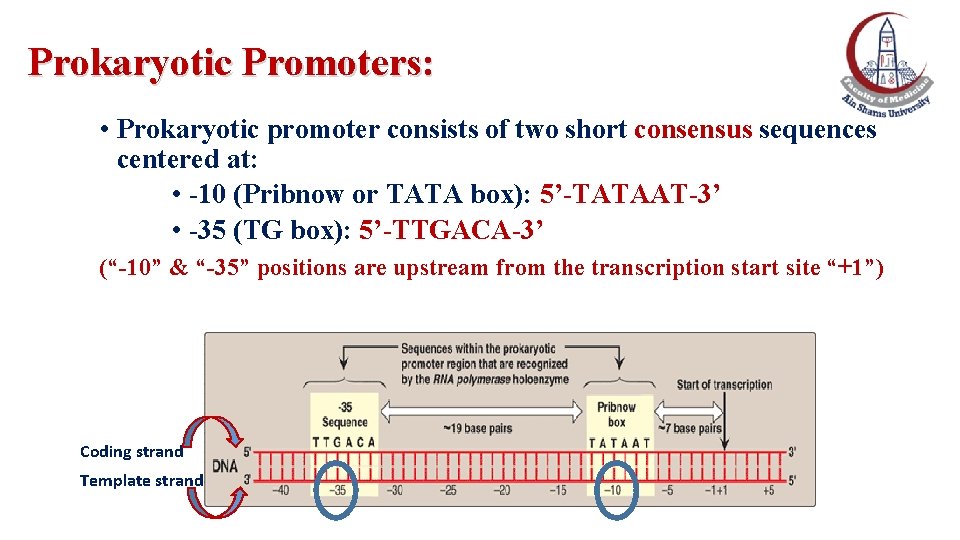
Prokaryotic Promoters: • Prokaryotic promoter consists of two short consensus sequences centered at: • -10 (Pribnow or TATA box): 5’-TATAAT-3’ • -35 (TG box): 5’-TTGACA-3’ (“-10” & “-35” positions are upstream from the transcription start site “+1”) Coding strand Template strand

Transcription Unit It is the segment of DNA between promoter and termination sequences and the product of transcription is called the primary transcript.

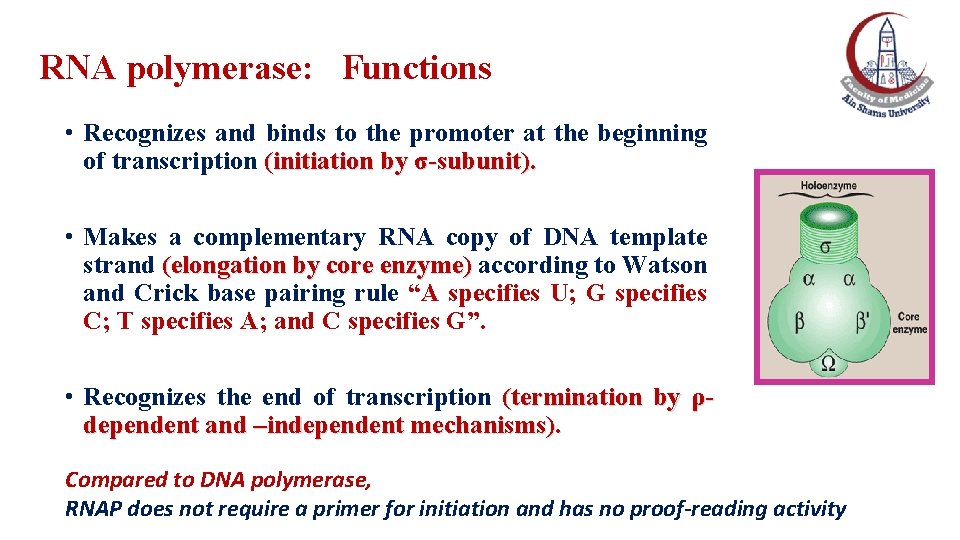
RNA polymerase: Functions • Recognizes and binds to the promoter at the beginning of transcription (initiation by σ-subunit). • Makes a complementary RNA copy of DNA template strand (elongation by core enzyme) according to Watson and Crick base pairing rule “A specifies U; G specifies C; T specifies A; and C specifies G”. • Recognizes the end of transcription (termination by ρdependent and –independent mechanisms). Compared to DNA polymerase, RNAP does not require a primer for initiation and has no proof-reading activity

Initiation

Transcription in Prokaryotes: The initiation Phase ØThe binding of RNA polymerase to DNA template results in local unwinding (melting) of the DNA helix. ØSupercoils can be relaxed by topoisomerase I and II. ØThen, RNAP begins to synthesize an RNA transcript (usually begins with purine). ØThe σ subunit is released after initiation of transcription.

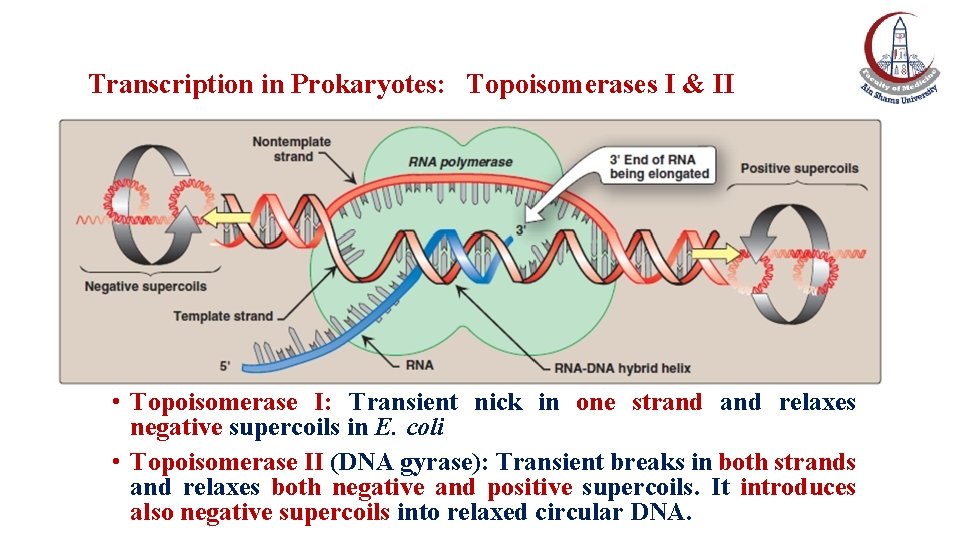
Transcription in Prokaryotes: Topoisomerases I & II • Topoisomerase I: Transient nick in one strand relaxes negative supercoils in E. coli • Topoisomerase II (DNA gyrase): Transient breaks in both strands and relaxes both negative and positive supercoils. It introduces also negative supercoils into relaxed circular DNA.

Elongation

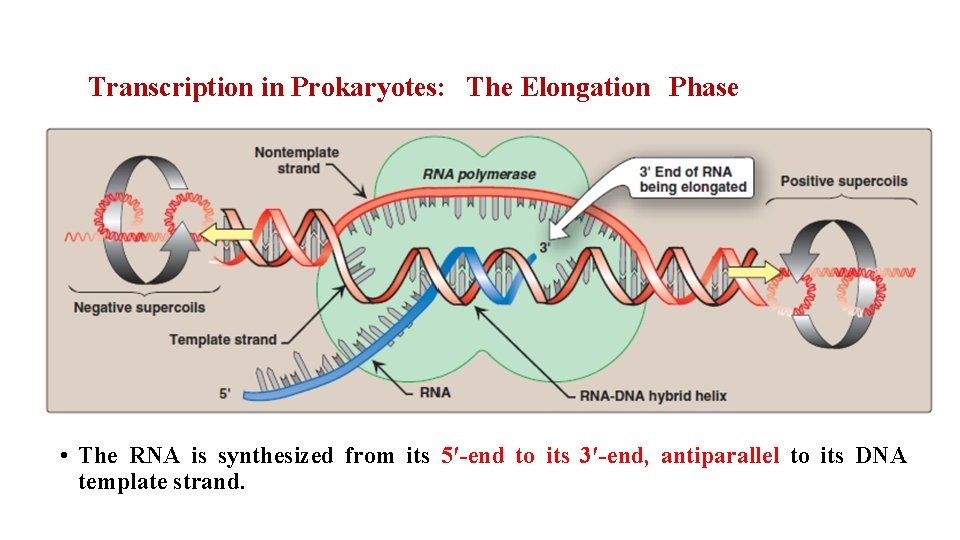
Transcription in Prokaryotes: The Elongation Phase The σ subunit is released after initiation of transcription and the core enzyme is able to leave the promoter and move along the DNA template strand in a processive manner (start of elongation phase).

Transcription in Prokaryotes: The Elongation Phase • The RNA is synthesized from its 5′-end to its 3′-end, antiparallel to its DNA template strand.

Termination

Transcription in Prokaryotes: the Termination Phase RNA polymerase recognizes a termination signal at the end of the DNA sequence to be transcribed (termination sequence). At this point, RNA polymerase stops transcription and releases the completed RNA molecule.

Termination mechanisms • Rho-independent transcription termination (Intrinsic termination) • Rho-dependent transcription termination (ATP dependent)

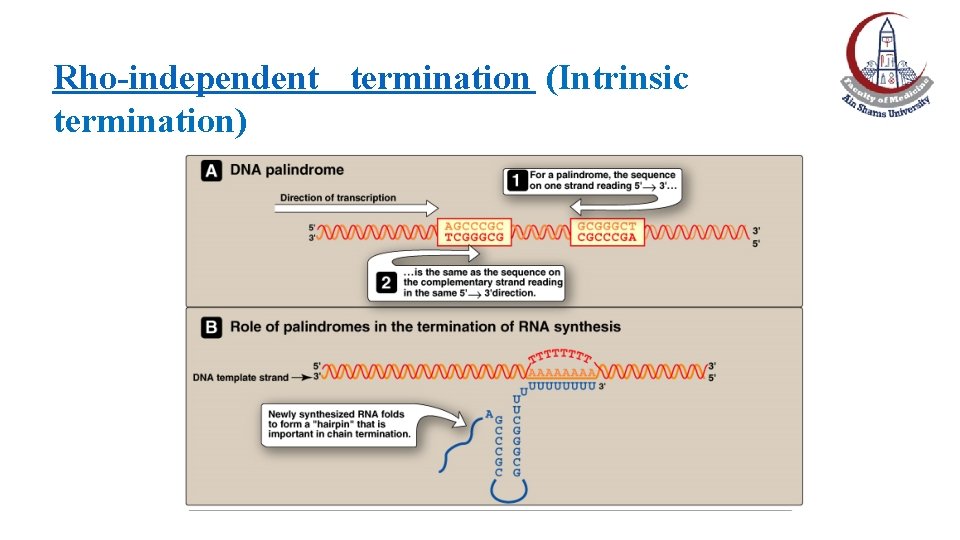
Rho-independent termination (Intrinsic termination) • A termination sequence is found in the DNA template that generates a self-complementary sequence in the newly transcribed RNA. • The GC-rich complementary bases join each other forming a stem and loop structure “hairpin” (folding of RNA) that leads to the dissociation of the RNAP from the DNA template and release of RNA. • Just beyond the hairpin, there is a string of Us at the 3` end of the RNA with the result of a weaker bonding with As of the DNA template and, hence, easier separation of RNA from its template.

Madam I’m Adam

Rho-independent termination (Intrinsic termination)

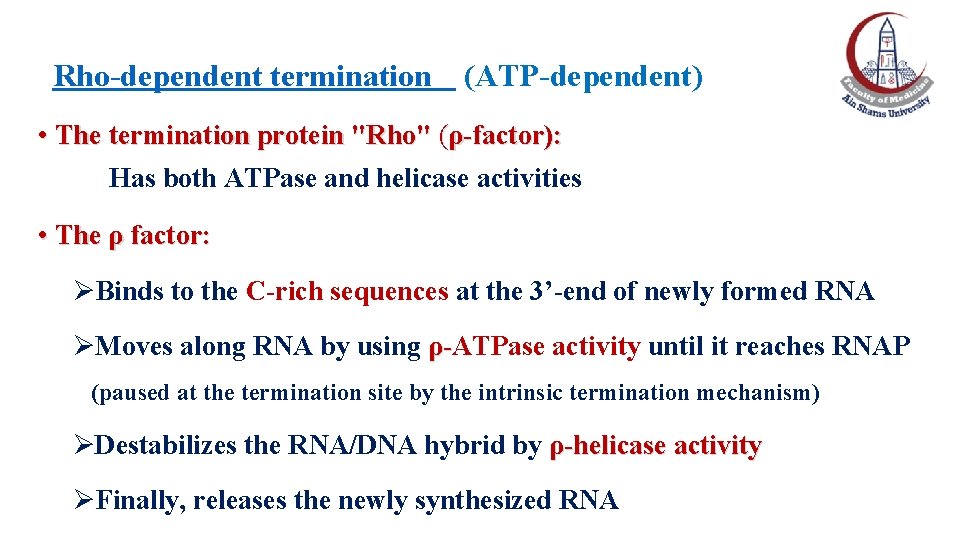
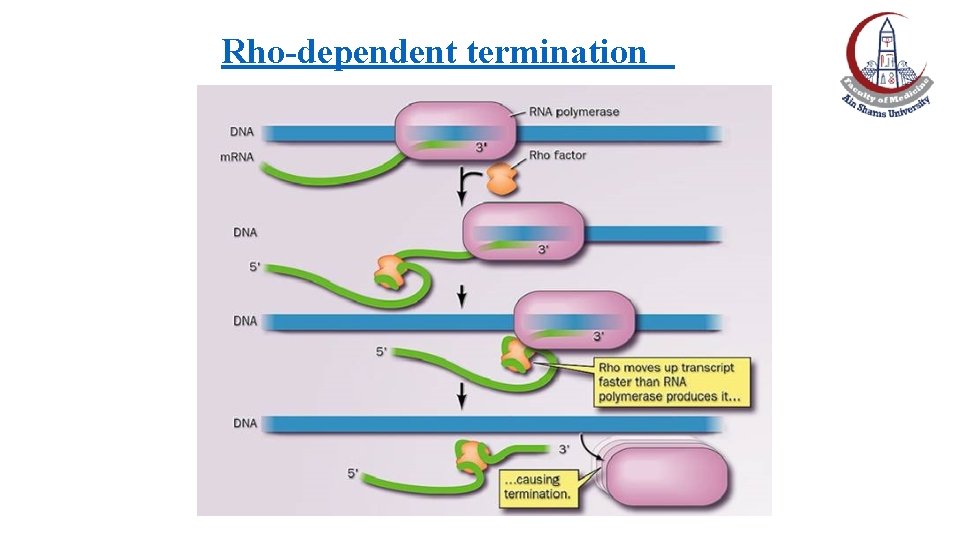
Rho-dependent termination (ATP-dependent) • The termination protein "Rho" (ρ-factor): Has both ATPase and helicase activities • The ρ factor: ØBinds to the C-rich sequences at the 3’-end of newly formed RNA ØMoves along RNA by using ρ-ATPase activity until it reaches RNAP ρ(paused at the termination site by the intrinsic termination mechanism) ØDestabilizes the RNA/DNA hybrid by ρ-helicase activity ØFinally, releases the newly synthesized RNA

Rho-dependent termination

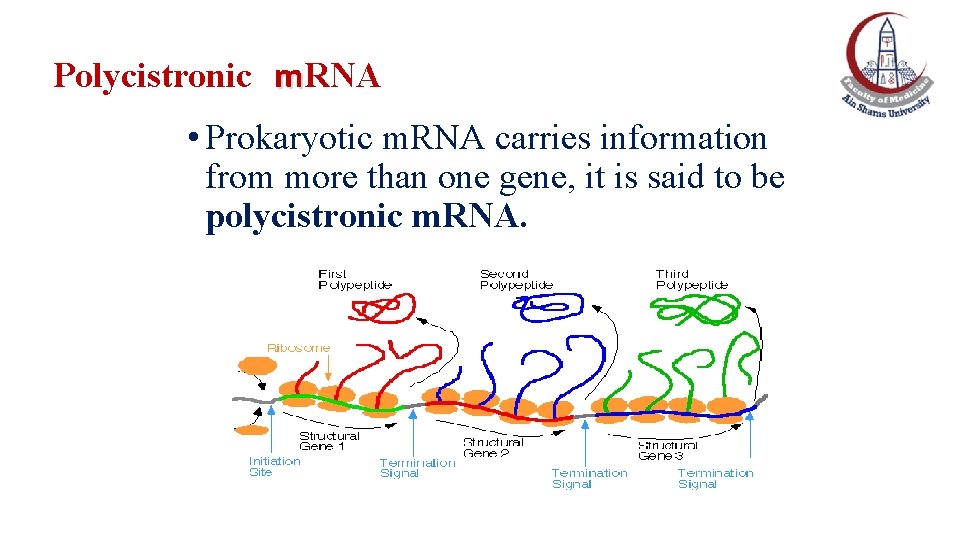
Polycistronic m. RNA • Prokaryotic m. RNA carries information from more than one gene, it is said to be polycistronic m. RNA.

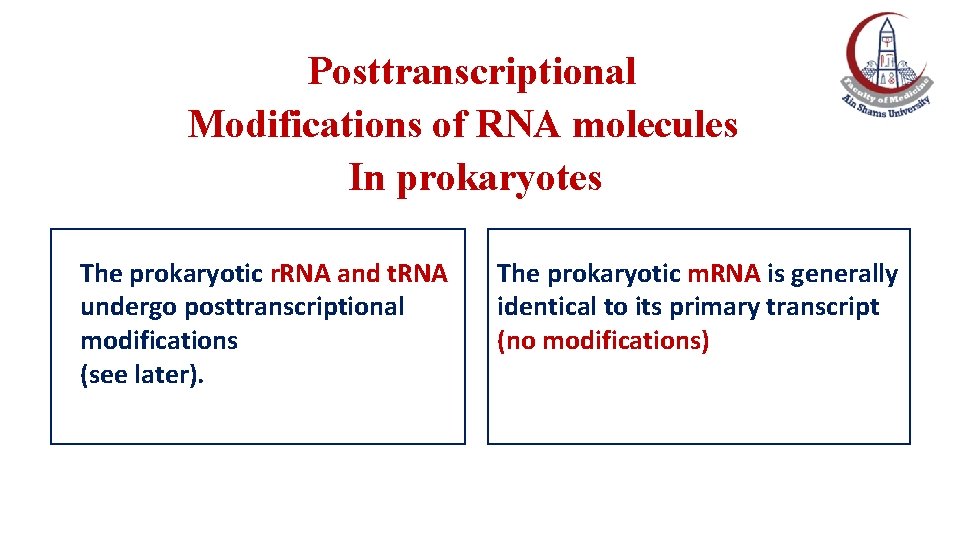
Posttranscriptional Modifications of RNA molecules In prokaryotes The prokaryotic r. RNA and t. RNA undergo posttranscriptional modifications (see later). The prokaryotic m. RNA is generally identical to its primary transcript (no modifications)

Transcription In eukaryotes

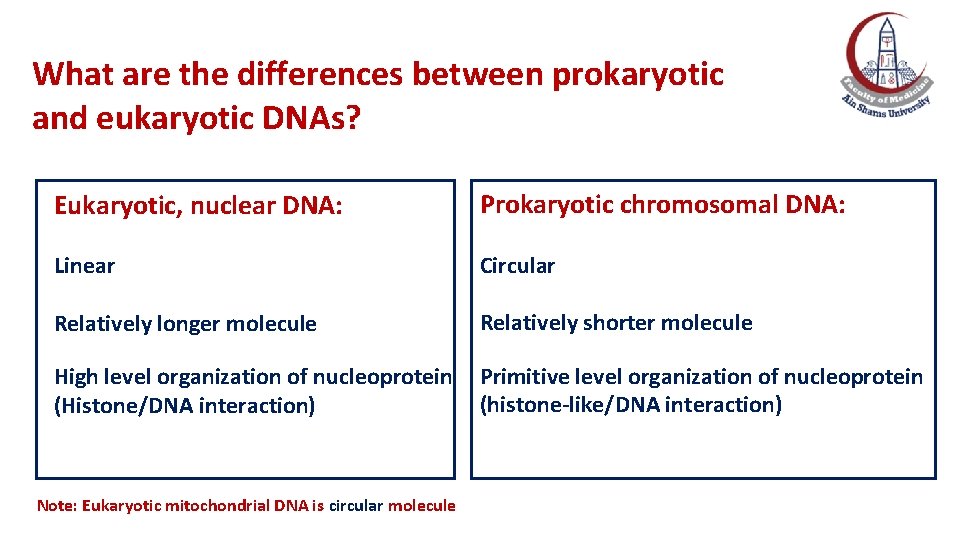
What are the differences between prokaryotic and eukaryotic DNAs? Eukaryotic, nuclear DNA: Prokaryotic chromosomal DNA: Linear Circular Relatively longer molecule Relatively shorter molecule High level organization of nucleoprotein (Histone/DNA interaction) Primitive level organization of nucleoprotein (histone-like/DNA interaction) Note: Eukaryotic mitochondrial DNA is circular molecule

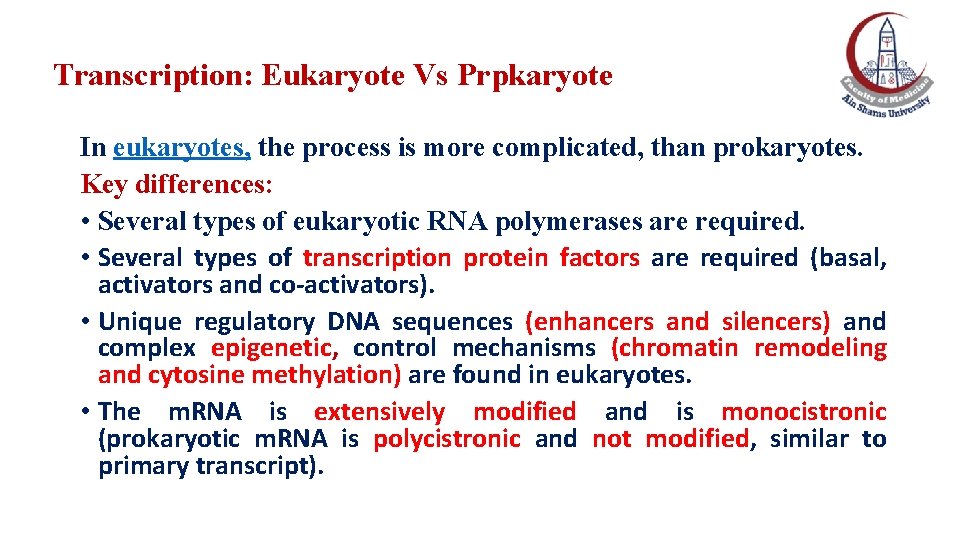
Transcription: Eukaryote Vs Prpkaryote In eukaryotes, the process is more complicated, than prokaryotes. Key differences: • Several types of eukaryotic RNA polymerases are required. • Several types of transcription protein factors are required (basal, activators and co-activators). • Unique regulatory DNA sequences (enhancers and silencers) and complex epigenetic, control mechanisms (chromatin remodeling and cytosine methylation) are found in eukaryotes. • The m. RNA is extensively modified and is monocistronic (prokaryotic m. RNA is polycistronic and not modified, similar to primary transcript).

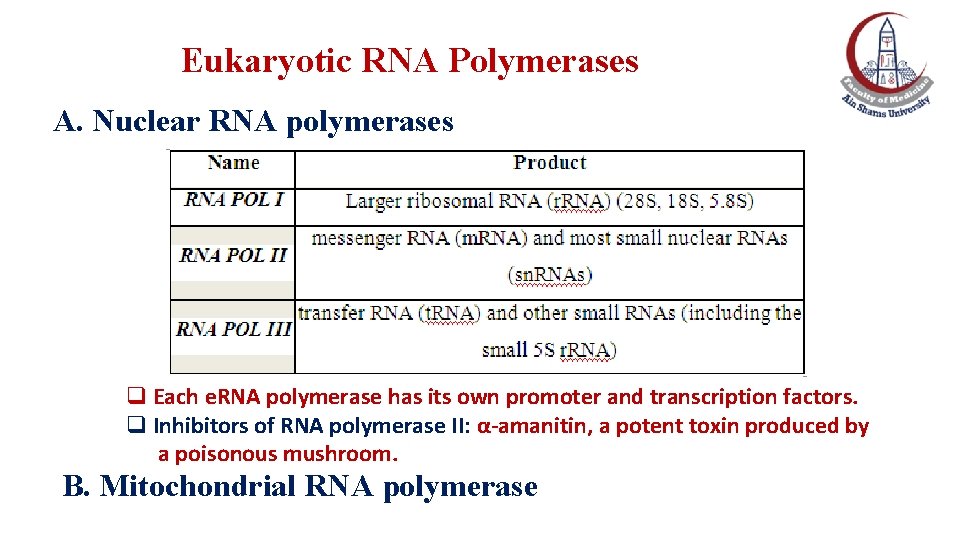
Eukaryotic RNA Polymerases A. Nuclear RNA polymerases q Each e. RNA polymerase has its own promoter and transcription factors. q Inhibitors of RNA polymerase II: α-amanitin, a potent toxin produced by a poisonous mushroom. B. Mitochondrial RNA polymerase

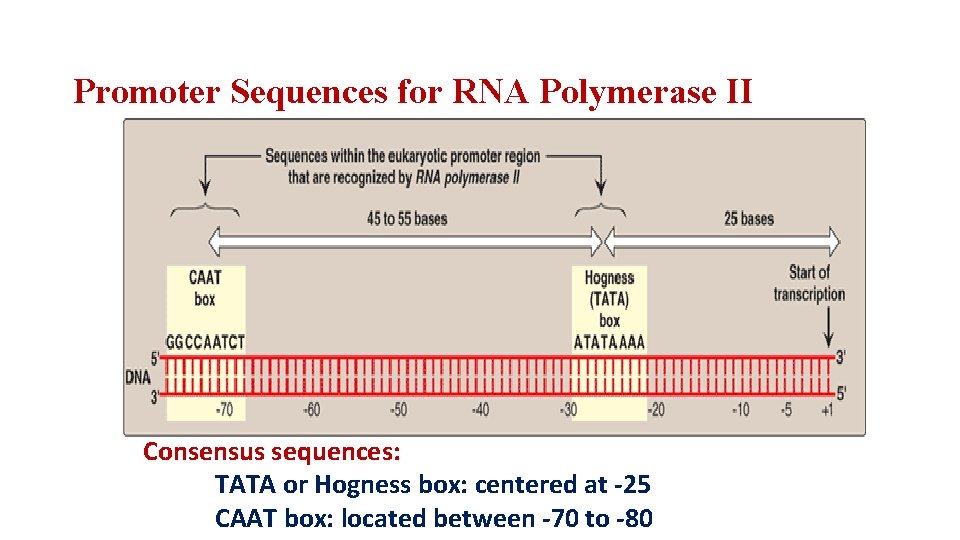
Promoter Sequences for RNA Polymerase II Consensus sequences: TATA or Hogness box: centered at -25 CAAT box: located between -70 to -80

RNA Polymerase II Promoter • Promoter sequences serve as binding sites for proteins known as transcription factors (TFs). • TFs interact with each others and recruit RNA polymerase II to the promoter region for initiation of transcription.

RNA Polymerase II Promoter • In contrast to holoenzyme of prokaryotes, e. RNA polymerase II does not itself recognize and bind to the promoter BUT it is recruited to the promoter by the general (or basal) TFs. • Initiation of transcription requires the binding of TFs to the regulatory DNA sequences (cis-acting elements) and also the interaction between these TFs and with e. RNA polymerase II. • Modulation of the efficiency of initiation process requires binding of specific TFs to other cis-acting regulatory sequences (enhancers/silencers) and also binding and interactions of these TFs with other proteins (coactivators). Slides 34 - 36, please refer to the lectures on regulation of gene expression for details

Transcription Factors Transcription factors (TFs) are proteins that are required for transcription in eukaryotes. TFs have DNA-binding domain (for binding to DNA) and protein-binding domain (for binding and interaction with RNAP and other proteins. TFs are trans-acting elements because they are encoded by other genes than the gene to be transcribed and are synthesized in the ribosomes. Then, TFs migrate to the nucleus for regulation of transcription.

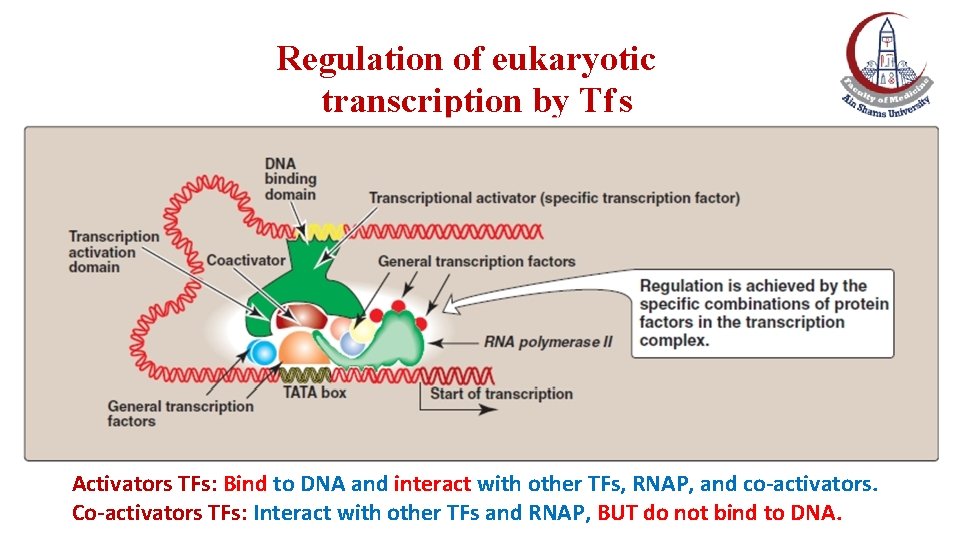
Regulation of eukaryotic transcription by Tf s Activators TFs: Bind to DNA and interact with other TFs, RNAP, and co-activators. Co-activators TFs: Interact with other TFs and RNAP, BUT do not bind to DNA.

Posttranscriptional Modifications of rna Intended Learning Outcomes 1. Identify posttranscriptional modifications of r. RNA and t. RNA in both prokaryotes and eukaryotes. 2. Describe posttranscriptional modifications of m. RNA in eukaryotes only.

Posttranscriptional Modifications of Prokaryotic and eukaryotic r. RNA

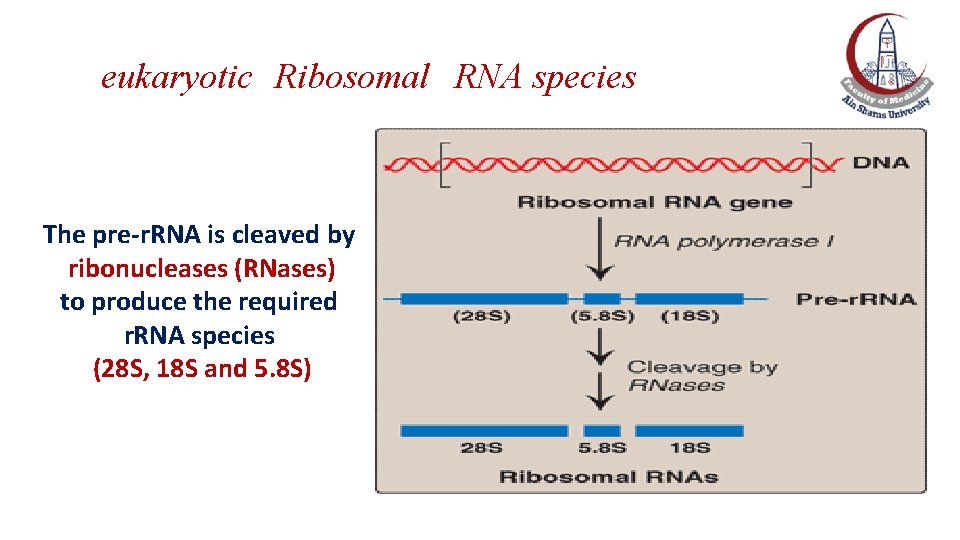
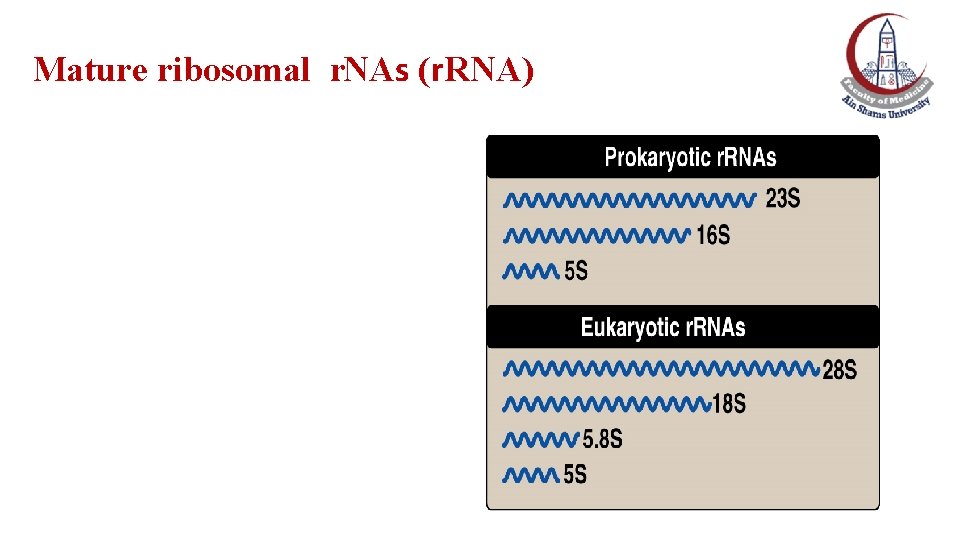
The pre-ribosomal RNA s The r. RNA of both prokaryotic and eukaryotic cells are synthesized from long precursor molecules called pre-ribosomal RNAs. The 23 S, 16 S and 5 S r. RNA of prokaryotes are synthesized by RNA polymerase from a single RNA precursor molecule. The 28 S, 18 S and 5. 8 S r. RNA of eukaryotes are synthesized by e. RNA polymerase I from a single precursor molecule. The 5 S r. RNA of eukaryotes is synthesized by e. RNA polymerase III and modified separately.

eukaryotic Ribosomal RNA species The pre-r. RNA is cleaved by ribonucleases (RNases) to produce the required r. RNA species (28 S, 18 S and 5. 8 S)

Mature ribosomal r. NAs (r. RNA)

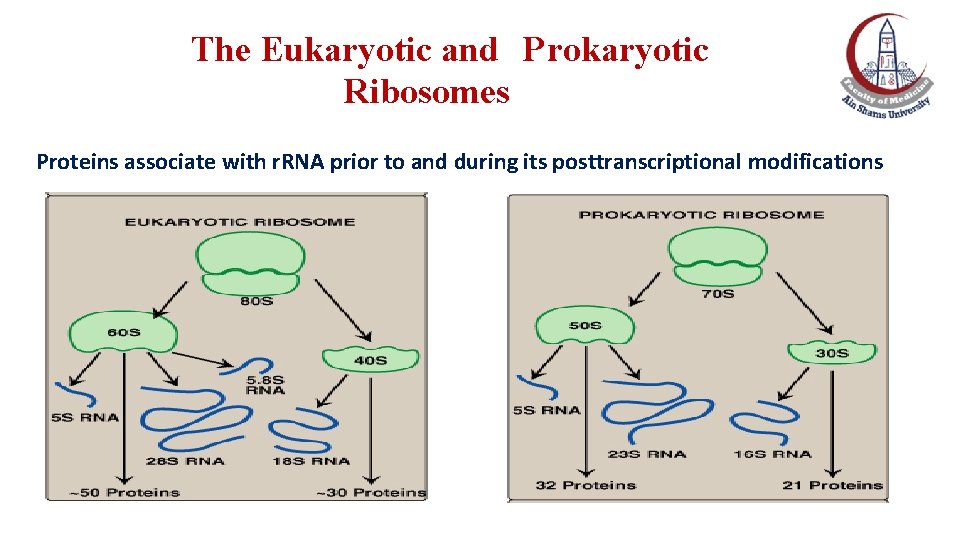
The Eukaryotic and Prokaryotic Ribosomes Proteins associate with r. RNA prior to and during its posttranscriptional modifications

Posttranscriptional Modifications of Prokaryotic and eukaryotic t. RNA

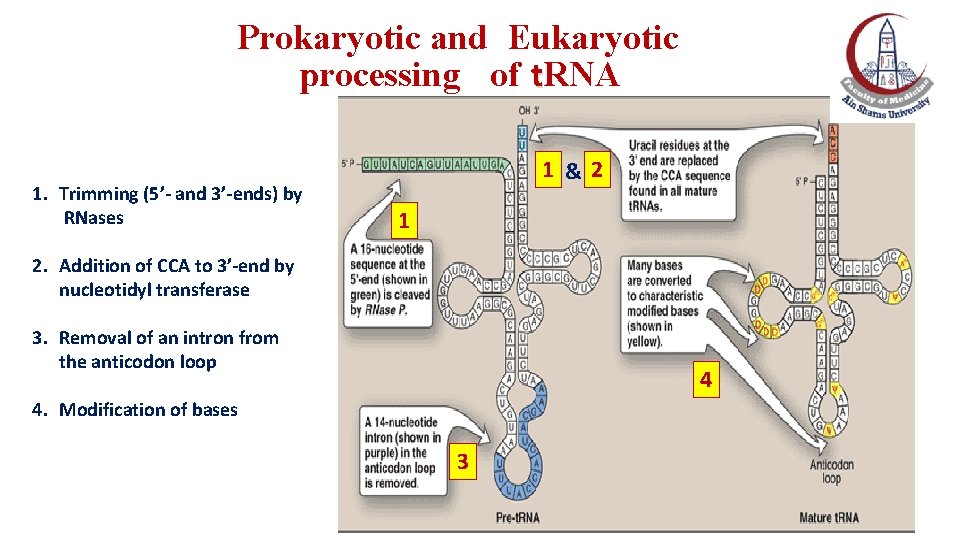
Prokaryotic and Eukaryotic processing of t. RNA 1. Trimming (5’- and 3’-ends) by RNases 1 &2 1 2. Addition of CCA to 3’-end by nucleotidyl transferase 3. Removal of an intron from the anticodon loop 4 4. Modification of bases 3

Prokaryotic and Eukaryotic processing of t. RNA 1. Similar to r. RNA, t. RNA is posttranscriptionally modified by cleavage of the original, longer precursor by ribonucleases: ØBoth the 5′- and the 3′-ends of the molecule must be trimmed. 2. Addition of a CCA by nucleotidyltransferase to the 3′-terminal end of t. RNA. 3. An intron must be removed from the anticodon loop. 3. Modification of bases at specific positions to produce “unusual bases”: e. g. , dihydrouracil, pseudouracil, thymine and methylated bases.

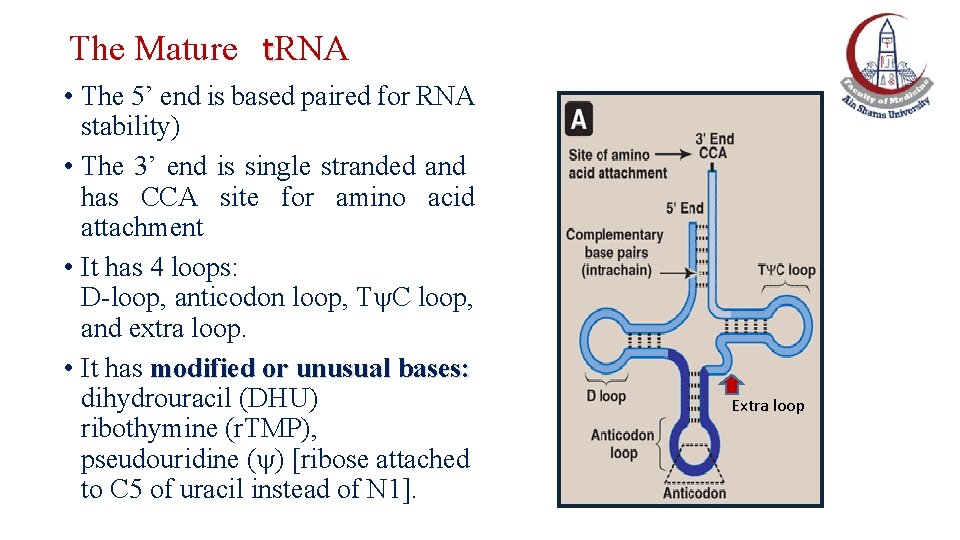
The Mature t. RNA • The 5’ end is based paired for RNA stability) • The 3’ end is single stranded and has CCA site for amino acid attachment • It has 4 loops: D-loop, anticodon loop, T C loop, and extra loop. • It has modified or unusual bases: dihydrouracil (DHU) ribothymine (r. TMP), pseudouridine ( ) [ribose attached to C 5 of uracil instead of N 1]. Extra loop

Posttranscriptional Modifications of Eukaryotic m. RNA

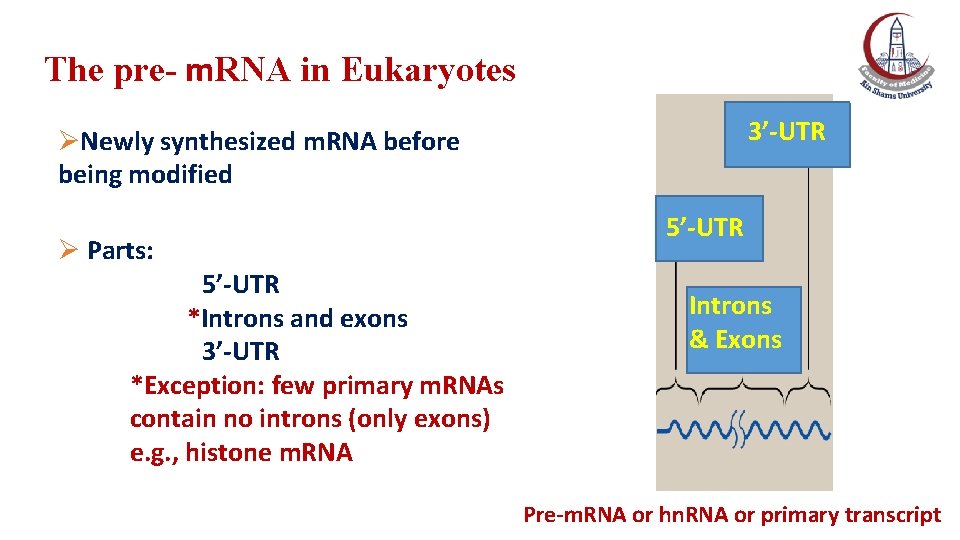
The pre- m. RNA in Eukaryotes 3’-UTR ØNewly synthesized m. RNA before being modified Ø Parts: 5’-UTR *Introns and exons 3’-UTR *Exception: few primary m. RNAs contain no introns (only exons) e. g. , histone m. RNA 5’-UTR Introns & Exons Pre-m. RNA or hn. RNA or primary transcript

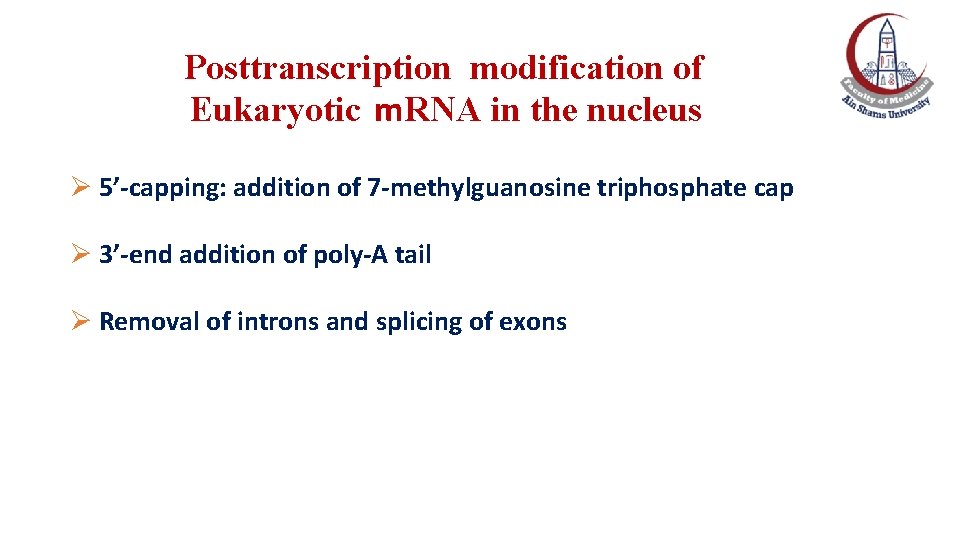
Posttranscription modification of Eukaryotic m. RNA in the nucleus Ø 5’-capping: addition of 7 -methylguanosine triphosphate cap Ø 3’-end addition of poly-A tail Ø Removal of introns and splicing of exons

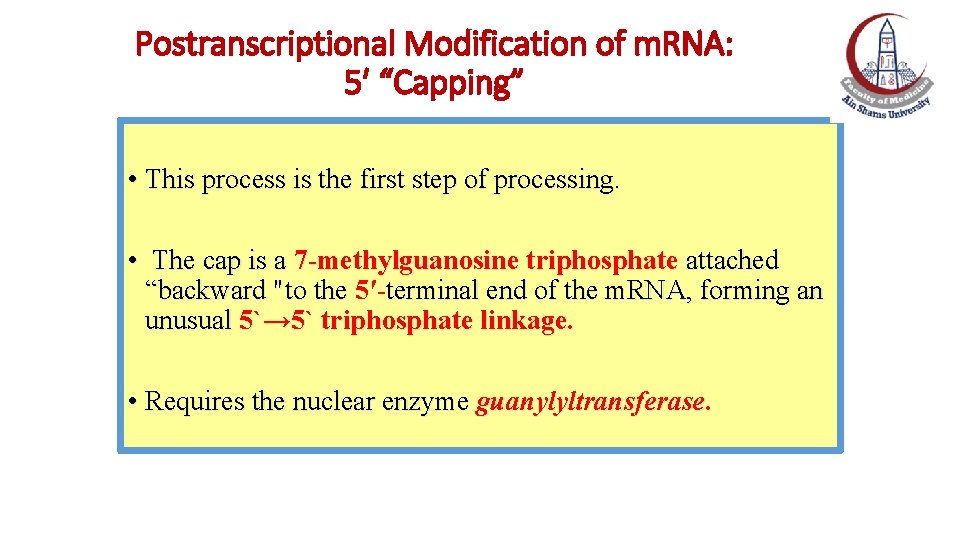
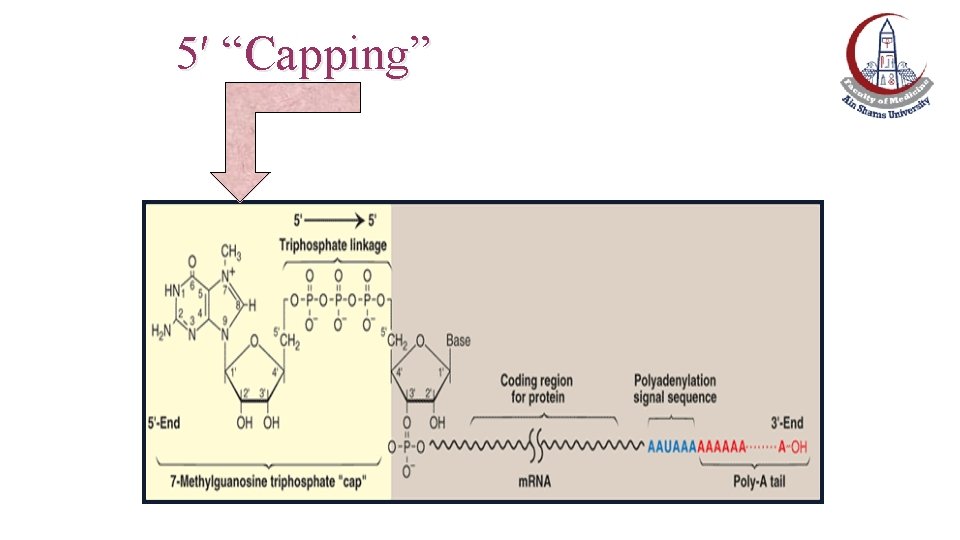
Postranscriptional Modification of m. RNA: 5′ “Capping” • This process is the first step of processing. • The cap is a 7 -methylguanosine triphosphate attached “backward "to the 5′-terminal end of the m. RNA, forming an unusual 5`→ 5` triphosphate linkage. • Requires the nuclear enzyme guanylyltransferase.

5′ “Capping”

Why? 5′ “Capping” • The addition of 7 -methylguanosine “cap” permits the initiation of translation, and helps stabilize the m. RNA. • Eukaryotic m. RNA lacking the cap are not efficiently translated.

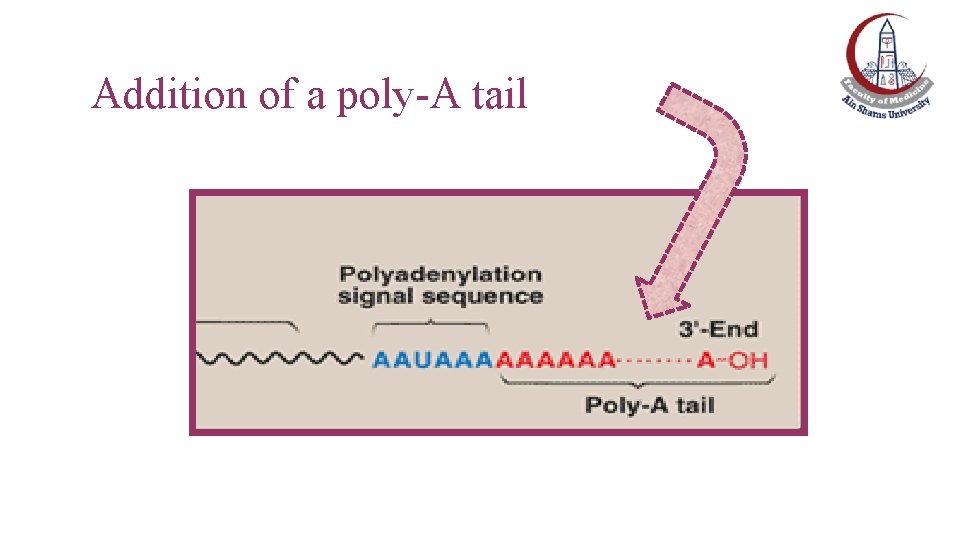
Postranscriptional Modification of m. RNA: Addition of 3’-poly-A tail • Most eukaryotic m. RNA have a chain of 40– 200 adenine nucleotides attached to the 3′-end (downstream of a polyadenylation signal AAUAAA) • This poly-A tail is not transcribed from the DNA. • This poly-A tail is added after transcription by the nuclear enzyme, polyadenylate polymerase, using ATP as the substrate. Exception: m. RNA of histones and some interferons have no poly-A tail

Addition of a poly-A tail

Why? (poly-A tails) • These tails help to stabilize the m. RNA • Facilitate their exit from the nucleus. Note, after the m. RNA enters the cytosol, the poly-A tail is gradually shortened.

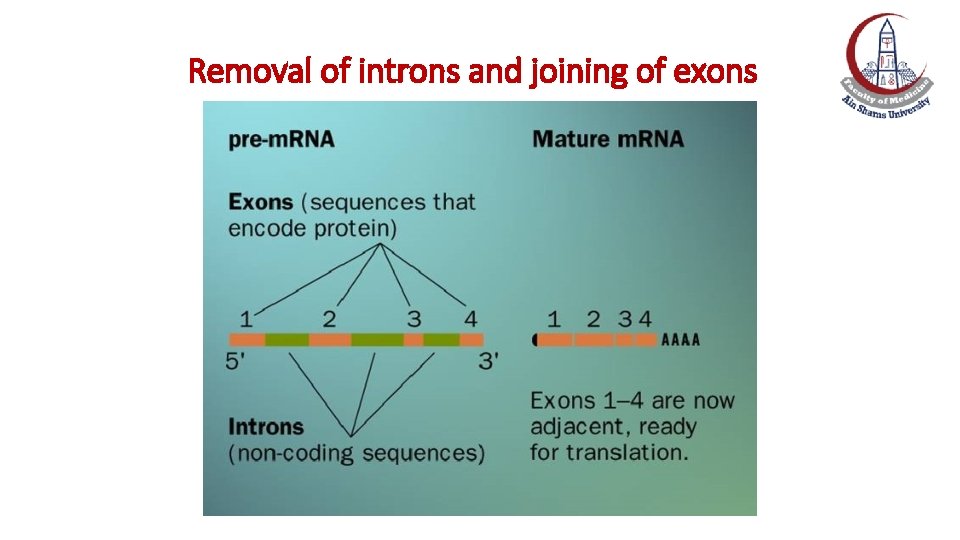
Postranscriptional Modification of m. RNA: Removal of Introns and Splicing of Exons • Maturation of eukaryotic m. RNA usually involves the removal of RNA sequences, which do not code for protein (introns, or intervening sequences) from the primary transcript. • The remaining coding sequences, the exons, are joined together (spliced) to form the mature m. RNA. Exception: m. RNA of histones has no introns

• Primary transcript of some genes can be as large as 1. 7 megabases (1, 700, 000 bases), while the mature (processed) m. RNA are under 10 kilobases (10, 000 bases)

Removal of introns and joining of exons

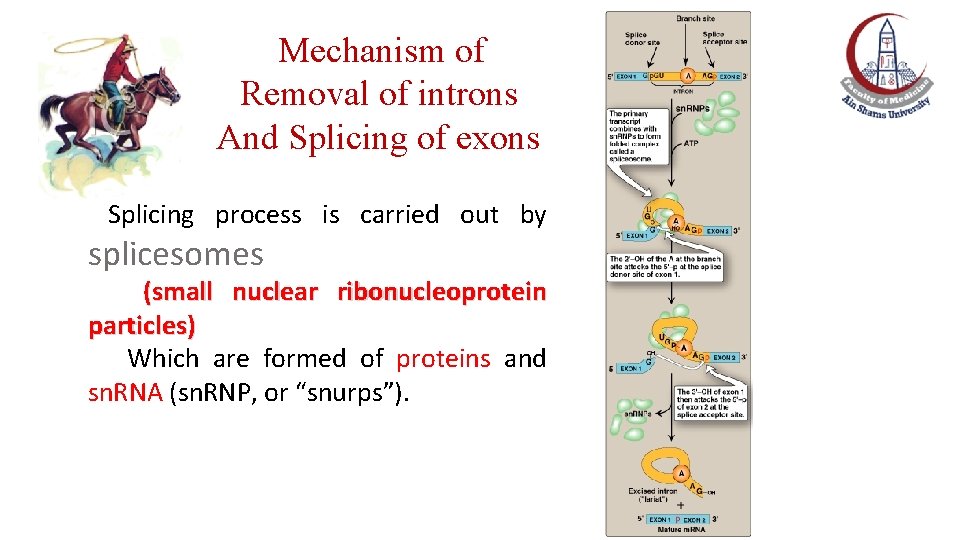
Mechanism of Removal of introns And Splicing of exons Splicing process is carried out by splicesomes (small nuclear ribonucleoprotein particles) Which are formed of proteins and sn. RNA (sn. RNP, or “snurps”).

Mechanism of Removal of introns And Splicing of exons Splicesomes (sn. RNP, or “snurps”) facilitate the removal of introns by forming base pairs with specific sequences at each end of the intron (lariat). Then, sn. RNP brings the sequences of neighboring exons into the correct alignment for splicing (formation of a phosphodiester bond between the two exons).

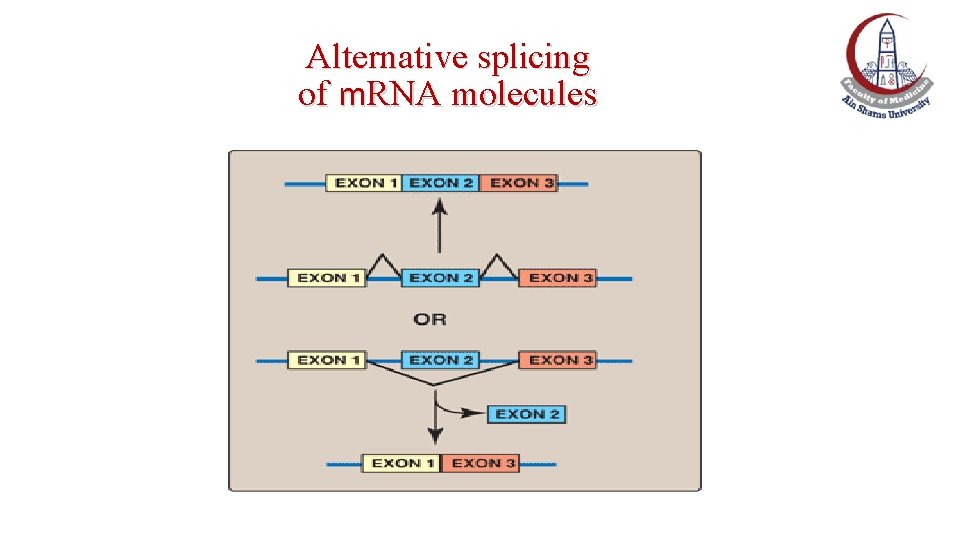
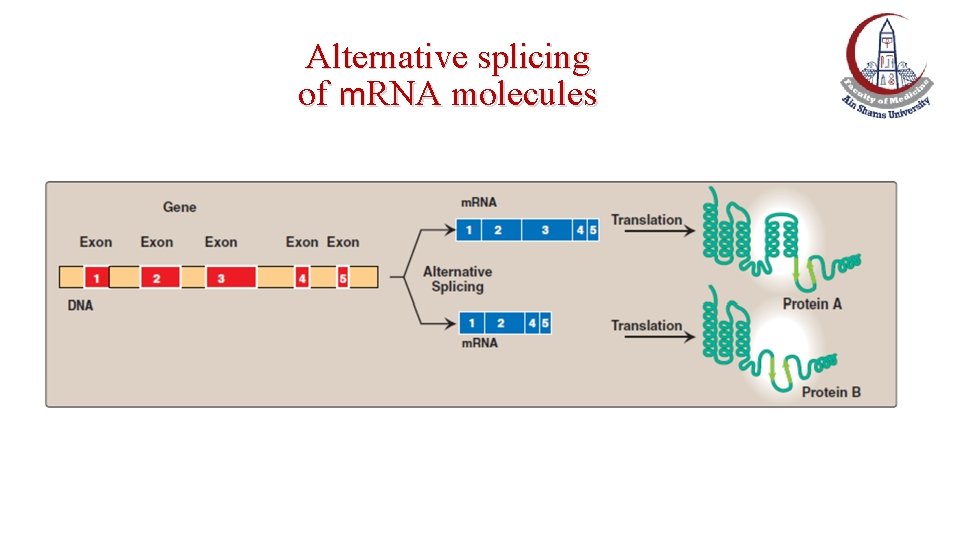
Alternative splicing of m. RNA molecules

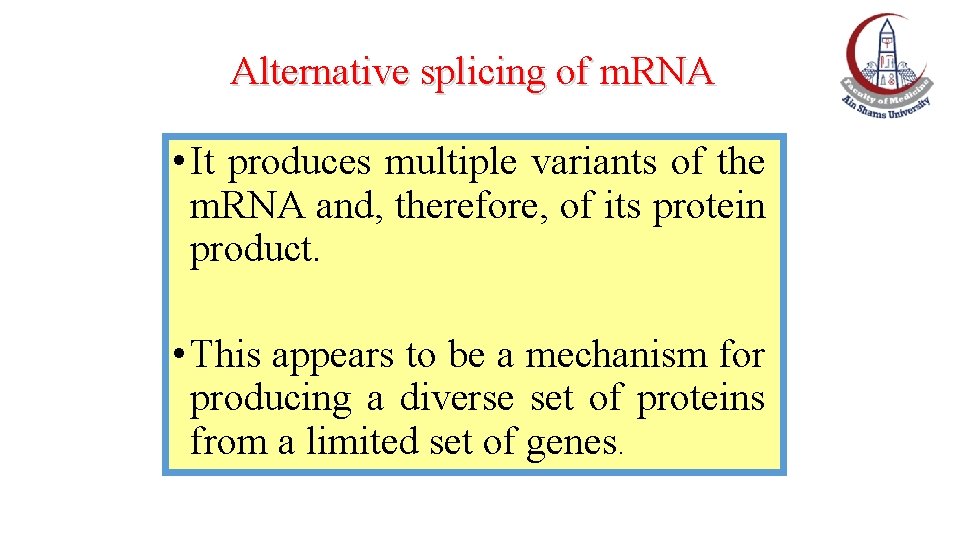
Alternative splicing of m. RNA • It produces multiple variants of the m. RNA and, therefore, of its protein product. • This appears to be a mechanism for producing a diverse set of proteins from a limited set of genes.

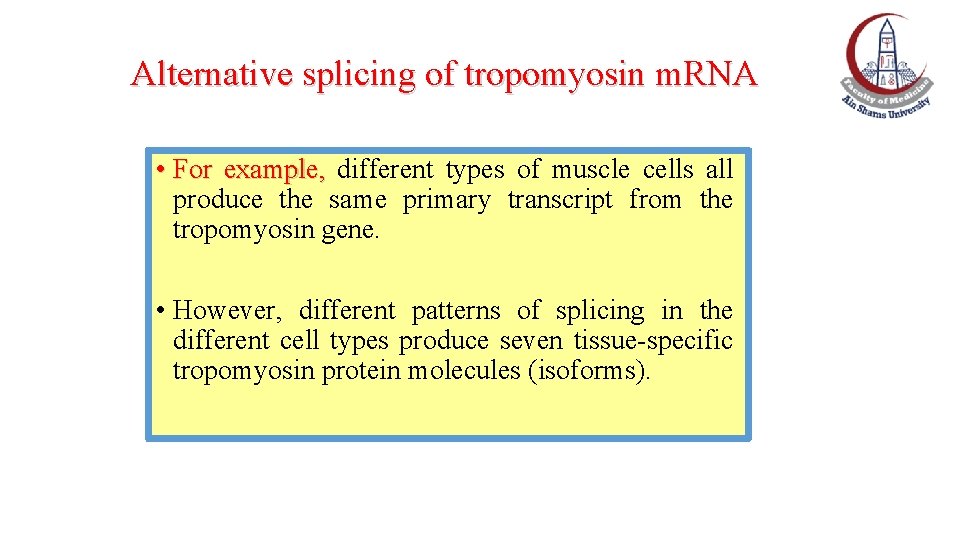
Alternative splicing of tropomyosin m. RNA • For example, different types of muscle cells all produce the same primary transcript from the tropomyosin gene. • However, different patterns of splicing in the different cell types produce seven tissue-specific tropomyosin protein molecules (isoforms).

Alternative splicing of m. RNA molecules

Clinical correlation Genetic diseases • Systemic lupus erythematosus: A fatal inflammatory disease, results from an autoimmune response in which the patient produces antibodies against self-proteins, including sn. RNP. • β-Thalassemia: A hemolytic disease caused by defective production of β-globin gene due to splice site mutation of its m. RNA.

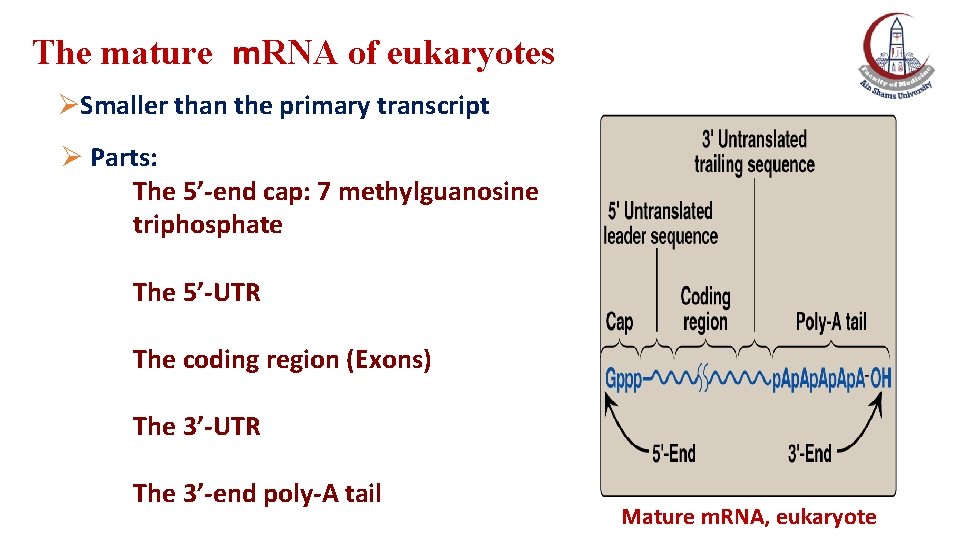
The mature m. RNA of eukaryotes ØSmaller than the primary transcript Ø Parts: The 5’-end cap: 7 methylguanosine triphosphate The 5’-UTR The coding region (Exons) The 3’-UTR The 3’-end poly-A tail Mature m. RNA, eukaryote

Reverse Transcriptase Reverse transcription: It is the process of synthesis of DNA from RNA by RNA-dependent DNA polymerase (reverse transcriptase). Retro viruses are viruses with RNA genome such as HIV (a virus causing AIDS), and H 1 N 1 (Swine flu virus) have the ability to transcribe RNA into DNA. Steps of reverse transcription • Following infection of the host cell, these viruses have a reverse transcriptase enzyme which uses viral RNA as a template for the synthesis of a complementary DNA strand. • The resulting DNA can be merged or integrated with the DNA genome of the host cell via an integrase enzyme causing the host cell to generate viral proteins.

Human Reverse Transcriptase e. g. , Telomerase Some eukaryotic cells (germ cells) contain an enzyme with reverse transcriptase activity, the telomerase, which lengthens the ends of linear chromosomes (telomeres).
