Towards an Integrated Physical Map of the Tomato





- Slides: 18

Towards an Integrated Physical Map of the Tomato Genome Mingsheng Chen Institute of Genetics and Developmental Biology Chinese Academy of Sciences January 13, 2008

l Reassemble the fingerprint data of Hind. III and Mbo. I libraries generated by Arizona Genomics Institute and Sanger l Manual editing and contig merging l Marker data generation: 3 D pool l FSD: Fingerprint Simulated Digest l Anchored contig information l Web. FPC

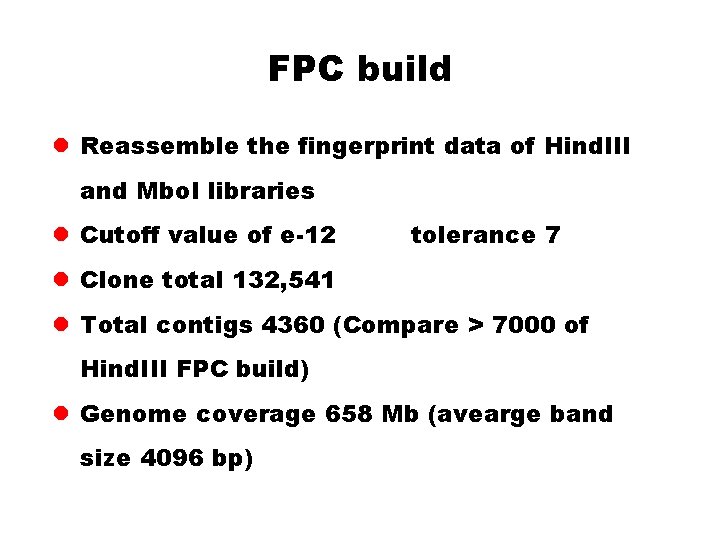
FPC build l Reassemble the fingerprint data of Hind. III and Mbo. I libraries l Cutoff value of e-12 tolerance 7 l Clone total 132, 541 l Total contigs 4360 (Compare > 7000 of Hind. III FPC build) l Genome coverage 658 Mb (avearge band size 4096 bp)

Manual Editing l Contig merging by BAC end searching combined with marker information l 4156 contigs (we only merged the contigs with anchoring information) l 837 markers (781 framework)

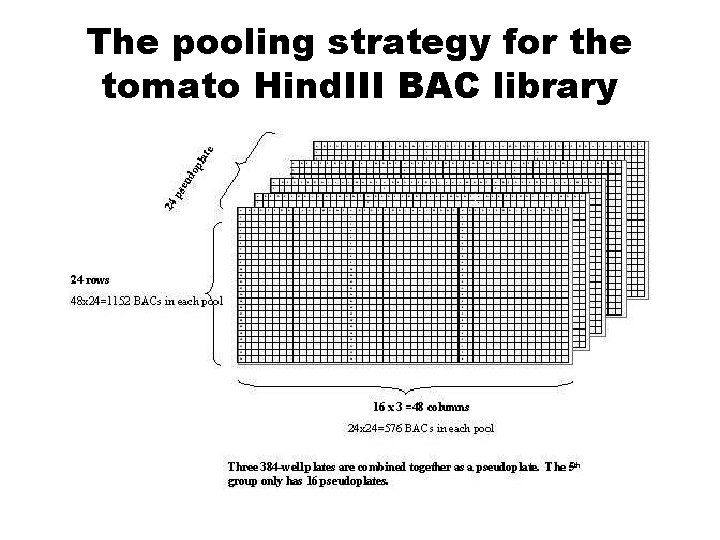
Marker l Overgo marker (provided by Ying Eileen Wang) l PCR-based approach: 3 -D pool

The pooling strategy for the tomato Hind. III BAC library

l 985 markers l 245 markers of chr 1/5/12 (Chen) l 249 markers of chr 3/7/9 (Li) l 247 markers of chr 4/6/10 (Ling) l 244 markers of chr 2/8/11 (Wang)

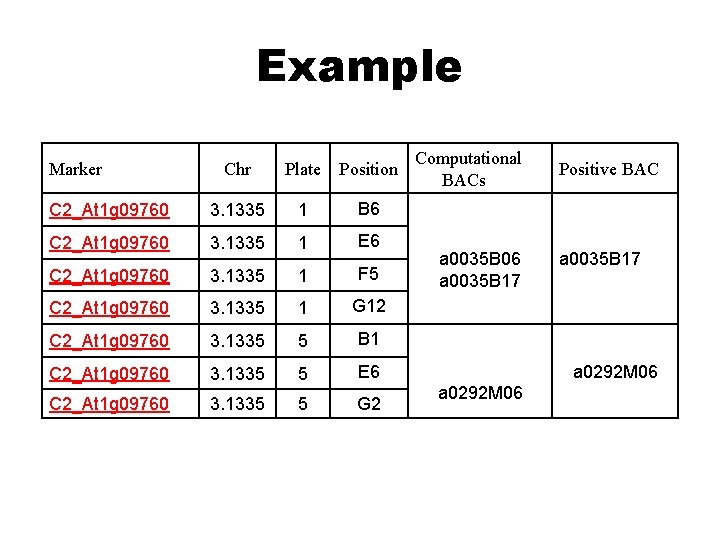
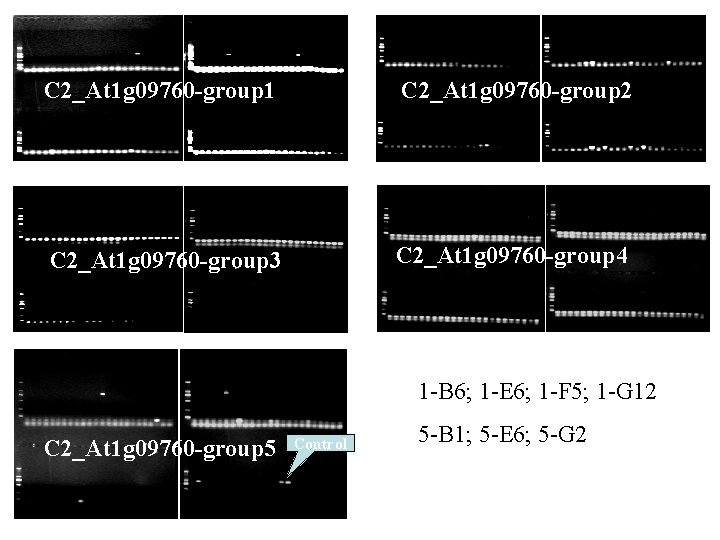
Example Marker Chr Plate Position C 2_At 1 g 09760 3. 1335 1 B 6 C 2_At 1 g 09760 3. 1335 1 E 6 C 2_At 1 g 09760 3. 1335 1 F 5 C 2_At 1 g 09760 3. 1335 1 G 12 C 2_At 1 g 09760 3. 1335 5 B 1 C 2_At 1 g 09760 3. 1335 5 E 6 C 2_At 1 g 09760 3. 1335 5 G 2 Computational BACs a 0035 B 06 a 0035 B 17 Positive BAC a 0035 B 17 a 0292 M 06

C 2_At 1 g 09760 -group 1 C 2_At 1 g 09760 -group 2 C 2_At 1 g 09760 -group 3 C 2_At 1 g 09760 -group 4 1 -B 6; 1 -E 6; 1 -F 5; 1 -G 12 C 2_At 1 g 09760 -group 5 Control 5 -B 1; 5 -E 6; 5 -G 2

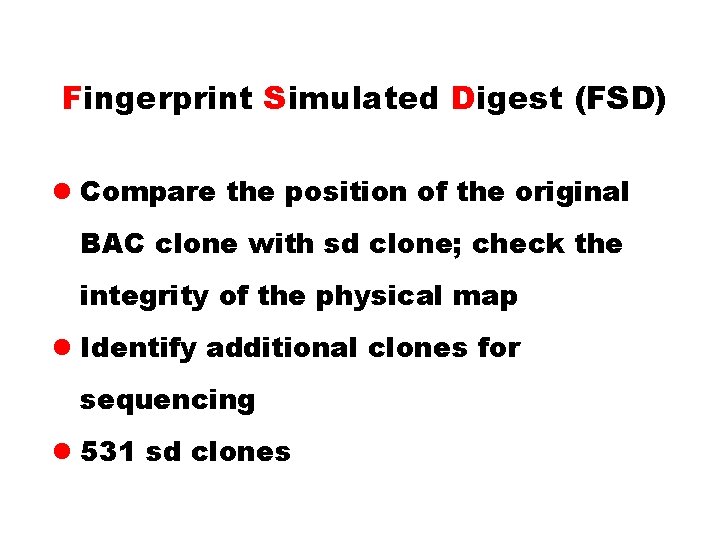
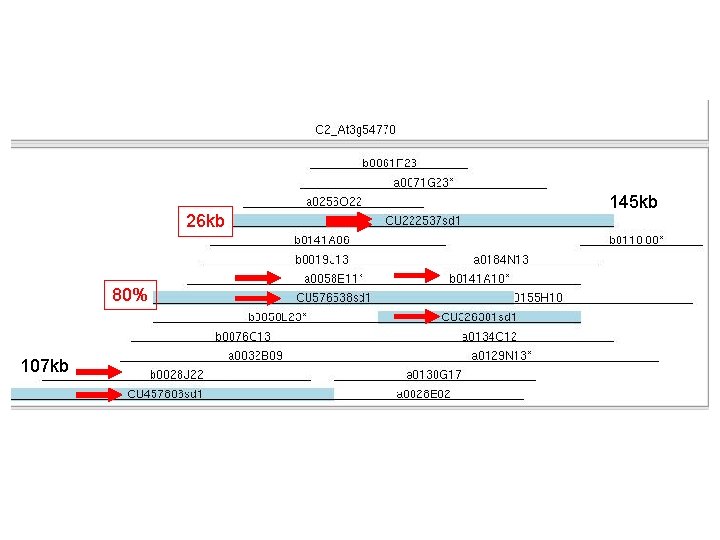
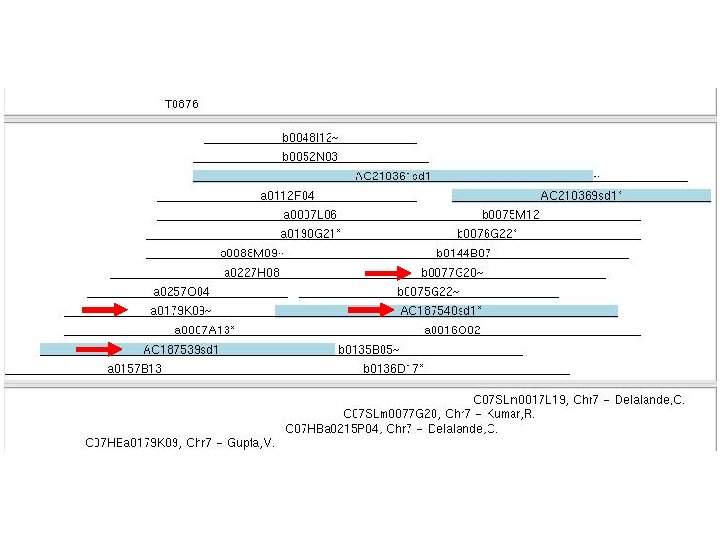
Fingerprint Simulated Digest (FSD) l Compare the position of the original BAC clone with sd clone; check the integrity of the physical map l Identify additional clones for sequencing l 531 sd clones

26 kb 80% 107 kb 145 kb


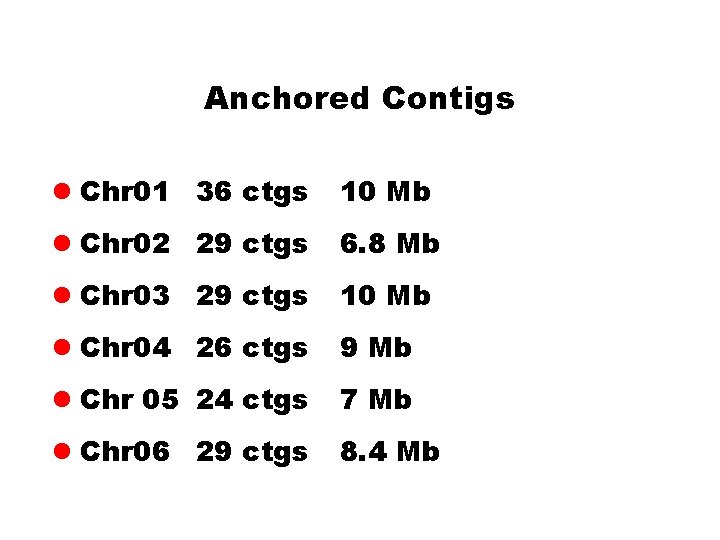
Anchored Contigs l Chr 01 36 ctgs 10 Mb l Chr 02 29 ctgs 6. 8 Mb l Chr 03 29 ctgs 10 Mb l Chr 04 26 ctgs 9 Mb l Chr 05 24 ctgs 7 Mb l Chr 06 29 ctgs 8. 4 Mb

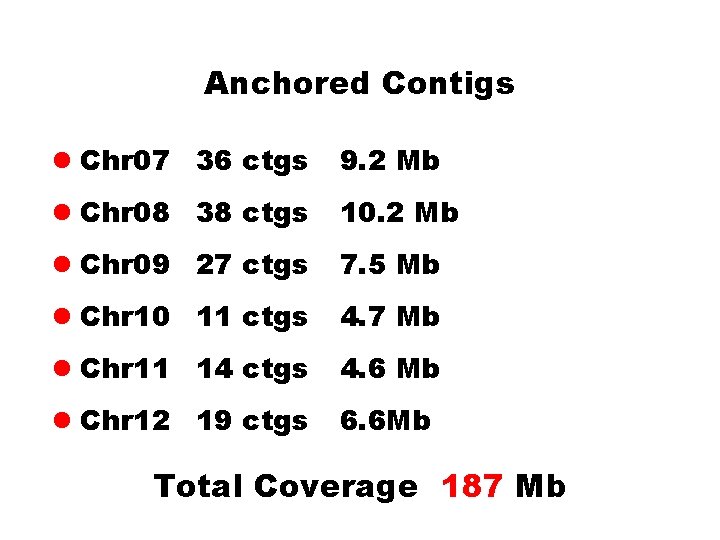
Anchored Contigs l Chr 07 36 ctgs 9. 2 Mb l Chr 08 38 ctgs 10. 2 Mb l Chr 09 27 ctgs 7. 5 Mb l Chr 10 11 ctgs 4. 7 Mb l Chr 11 14 ctgs 4. 6 Mb l Chr 12 19 ctgs 6. 6 Mb Total Coverage 187 Mb

Web. FPC http: //tomato. genetics. ac. cn/


