Topics in Integrative genomics and Proteomics studies http





























- Slides: 76

Topics in Integrative genomics and Proteomics studies 유전-단백체 통합 연구 http: //tcr. amegroups. com/article/view/10134/html

Course Introduction § Advances in genomic and proteomic technologies have led a huge increase in the generation of detail genomic and proteomic data more than ever. § There have been considerable efforts to disseminate and comprehend the data in the given biological and disease conditions. § The objective of this course is to introduce the emerging field of integrated genomics and proteomics (even metabolomics) studies in biomedical research. § The topics to be discussed the current developments of genome sequencing and proteome technologies. The course will also emphasize a special interest on novel annotation fields, such as that of phenotypes, and highlights the recent efforts focused on the integrating gene annotations in the disease context.

Course schedule • 9/03 -Overview • 9/10, 17, 10/01, 10/08 -Genomics (Dr. Eun. Goo Jung) • 10/15 - Midterm • 10/22, 29, 11/05, 11/12, 11/19, 11/26, 12/03 -Proteogenomics • 12/10 Final Exam

Proteogenomics Research: On the Frontier of Precision Medicine • https: //youtu. be/7 qp. KPi. OZrks

Learning Objectives • Define Genomics and Proteomics • Different applications of genomics and proteomics

Genomics vs Proteomics • Genomics : the systematic study of the entire genome of an organism o Physically map the genome arrangement(assign exact positions in the genome to the various genes/non-coding regions) o Human Genome Project § Began in 1990 , completed in 2003 § Total human genome size is in the region of 3. 2 gigabases (Gb) § Less than 1/3 of genome is transcribed into RNA and only 5% of that RNA is believed to encode polypeptides • Proteomics : direct examination of the expressed proteins in the cell • Systematic and comprehensive analysis of the proteins(proteome) expressed in the cell and their functions • Direct comparison of protein expression • Changes in cellular protein profiles with cellular conditions Genome sequences do not always provide a direct link to biological activity

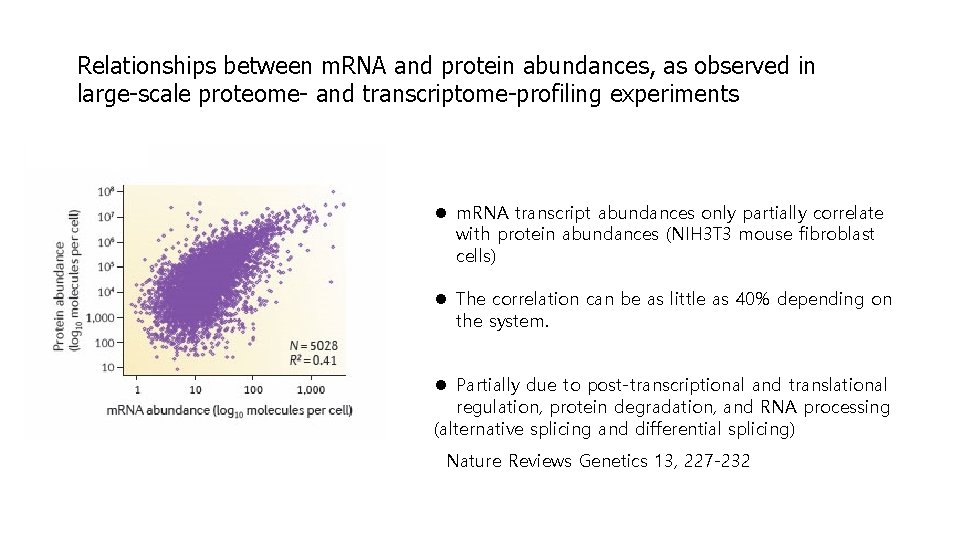
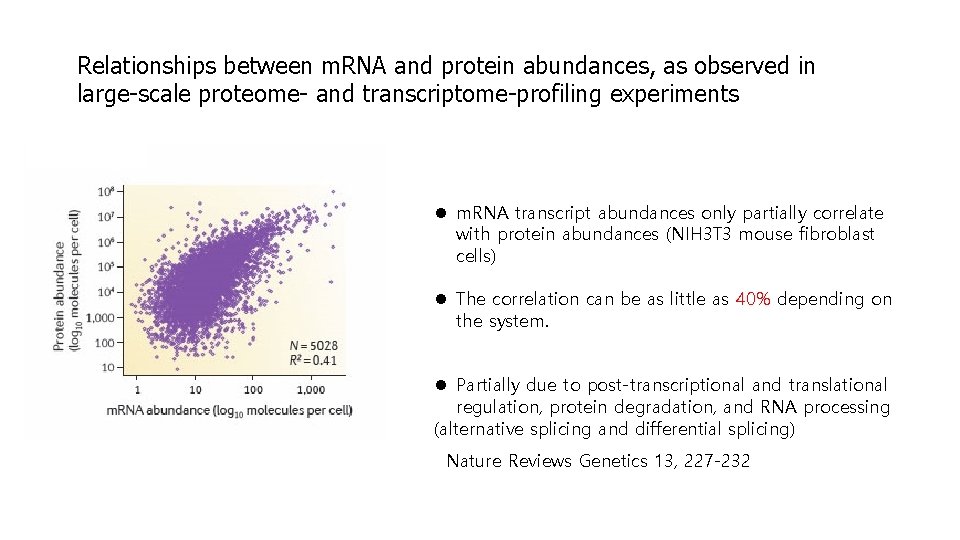
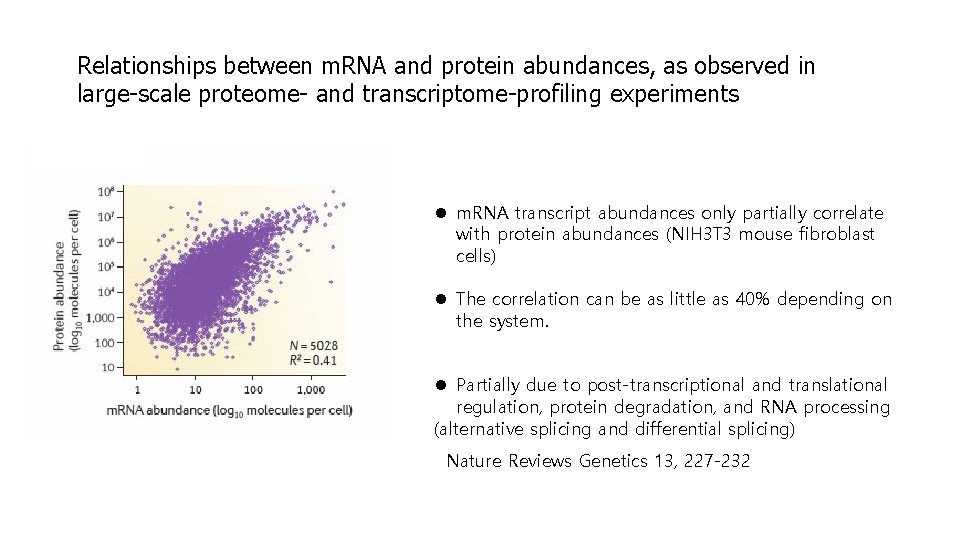
Relationships between m. RNA and protein abundances, as observed in large-scale proteome- and transcriptome-profiling experiments l m. RNA transcript abundances only partially correlate with protein abundances (NIH 3 T 3 mouse fibroblast cells) l The correlation can be as little as 40% depending on the system. l Partially due to post-transcriptional and translational regulation, protein degradation, and RNA processing (alternative splicing and differential splicing) Nature Reviews Genetics 13, 227 -232

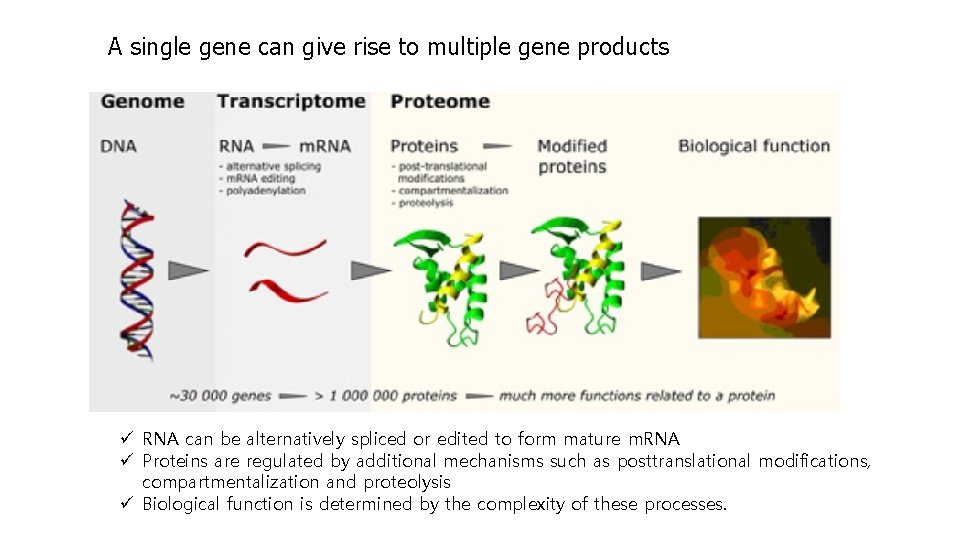
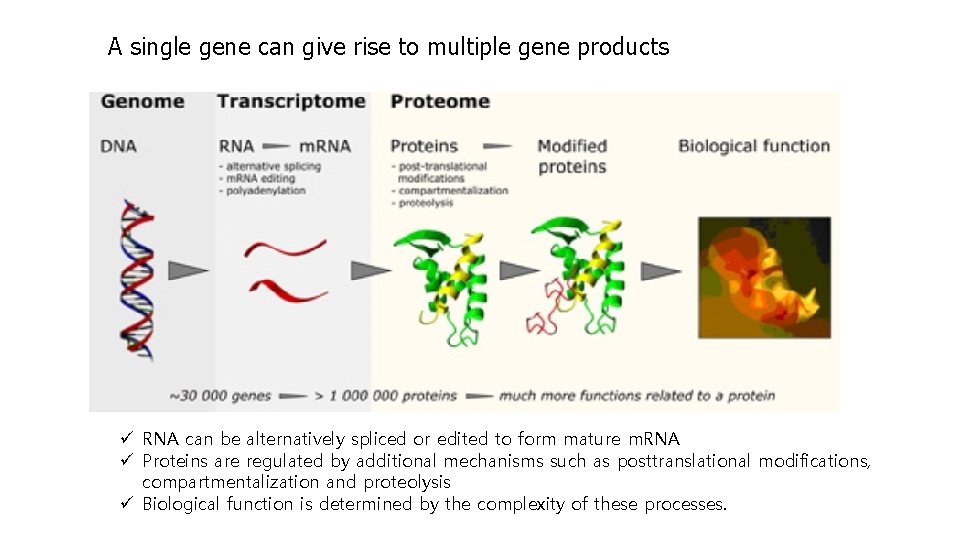
A single gene can give rise to multiple gene products ü RNA can be alternatively spliced or edited to form mature m. RNA ü Proteins are regulated by additional mechanisms such as posttranslational modifications, compartmentalization and proteolysis ü Biological function is determined by the complexity of these processes.

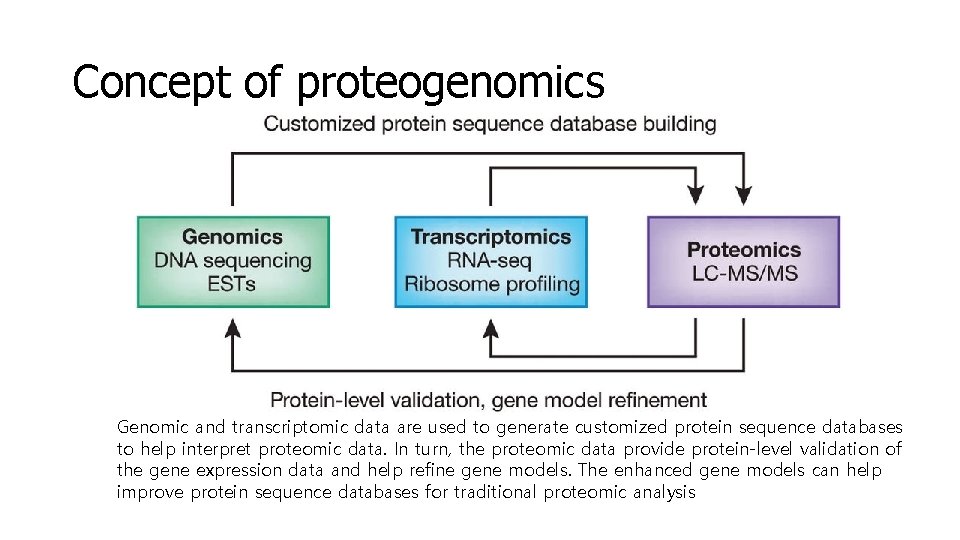
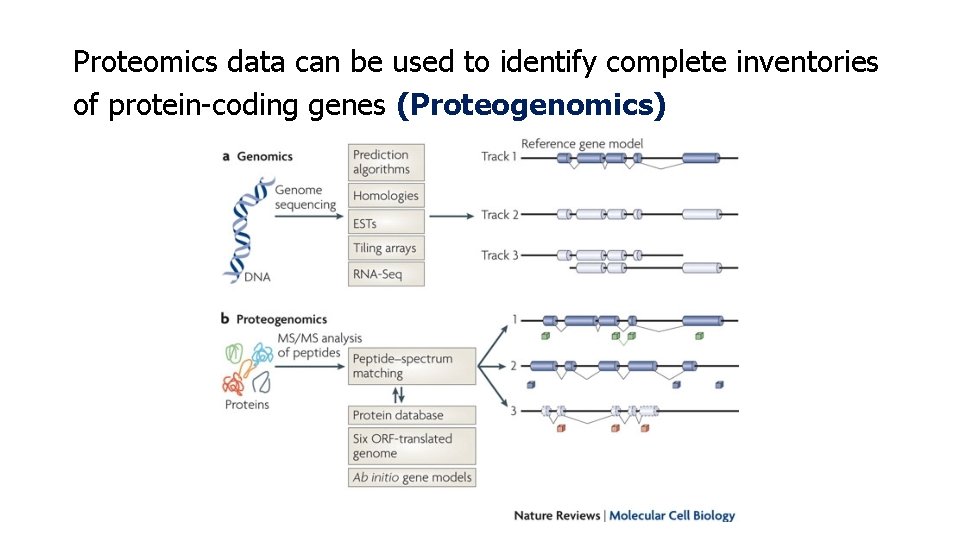
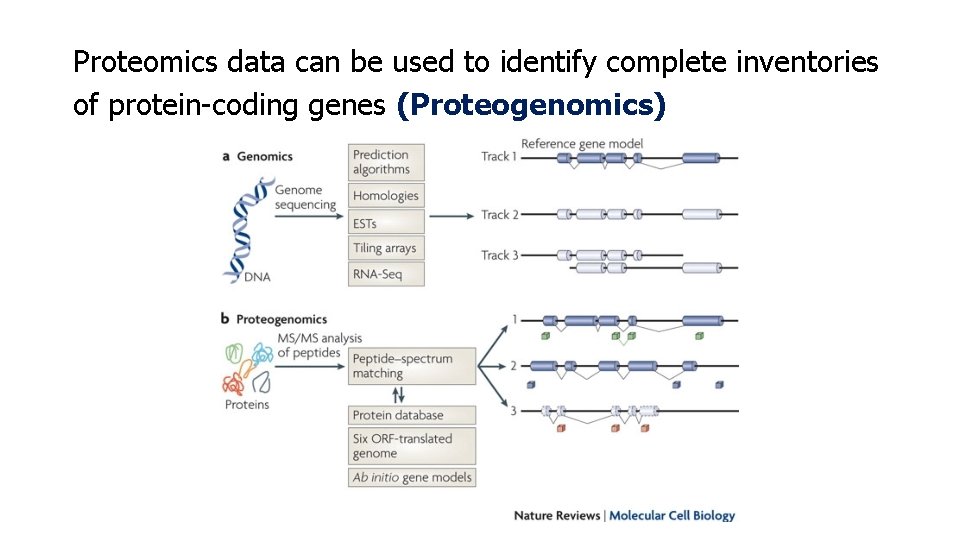
Concept of proteogenomics Genomic and transcriptomic data are used to generate customized protein sequence databases to help interpret proteomic data. In turn, the proteomic data provide protein-level validation of the gene expression data and help refine gene models. The enhanced gene models can help improve protein sequence databases for traditional proteomic analysis

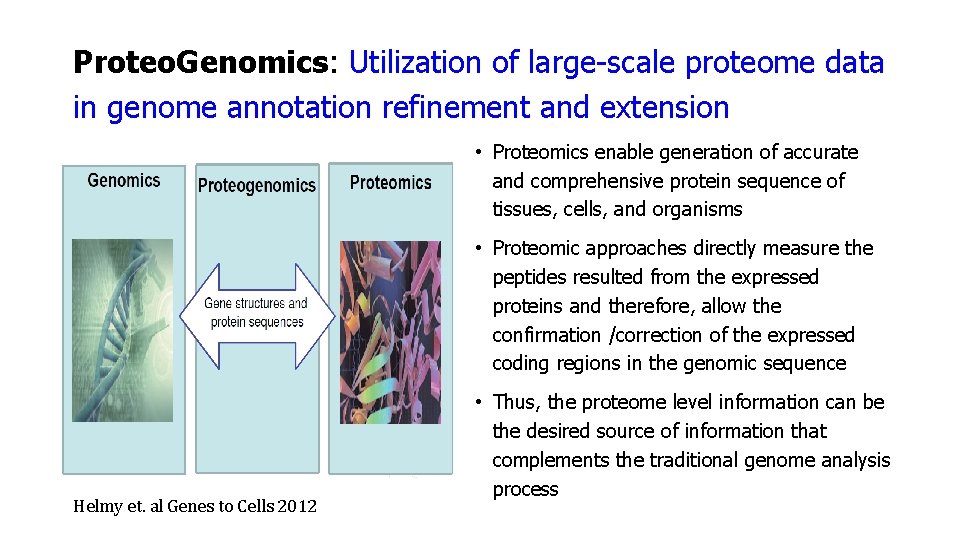
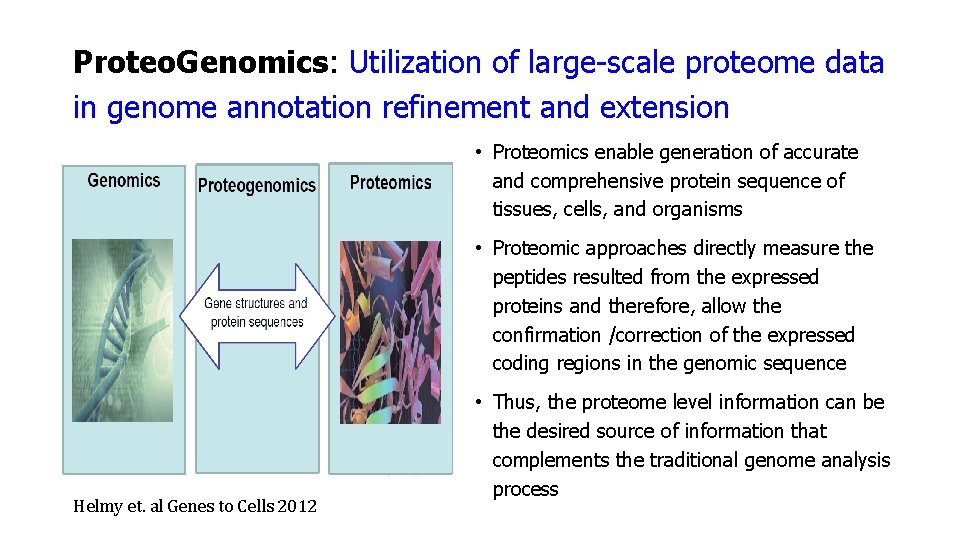
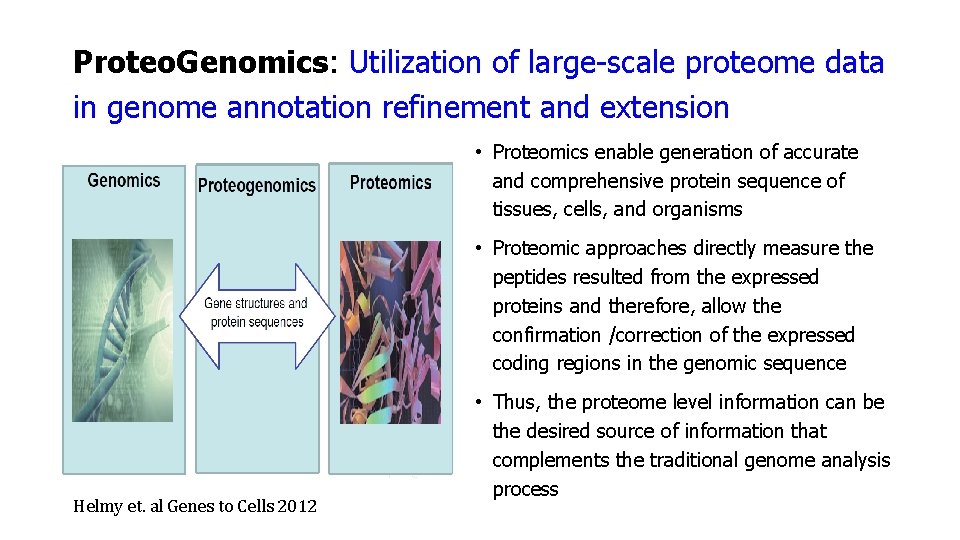
Proteo. Genomics: Utilization of large-scale proteome data in genome annotation refinement and extension • Proteomics enable generation of accurate and comprehensive protein sequence of tissues, cells, and organisms • Proteomic approaches directly measure the peptides resulted from the expressed proteins and therefore, allow the confirmation /correction of the expressed coding regions in the genomic sequence Helmy et. al Genes to Cells 2012 • Thus, the proteome level information can be the desired source of information that complements the traditional genome analysis process

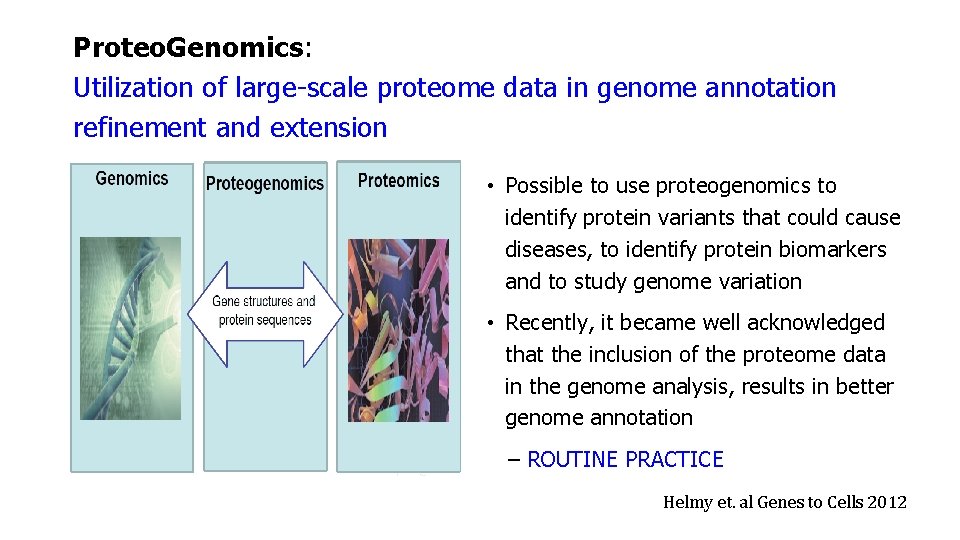
Proteo. Genomics: Utilization of large-scale proteome data in genome annotation refinement and extension • Possible to use proteogenomics to identify protein variants that could cause diseases, to identify protein biomarkers and to study genome variation • Recently, it became well acknowledged that the inclusion of the proteome data in the genome analysis, results in better genome annotation – ROUTINE PRACTICE Helmy et. al Genes to Cells 2012

Cancer Proteogenomics Cancer is characterized by altered expression of tumor drivers and suppressors • Results from gene mutations causing changes in protein expression, activity Cancer Proteomics • • Are genomic variants evident at the protein level? What is their effect on protein function associated cancers? What proteins have altered expression in tumor tissue? Can these proteins be used as cancer biomarkers for disease prevention, prognosis and targeted therapy?


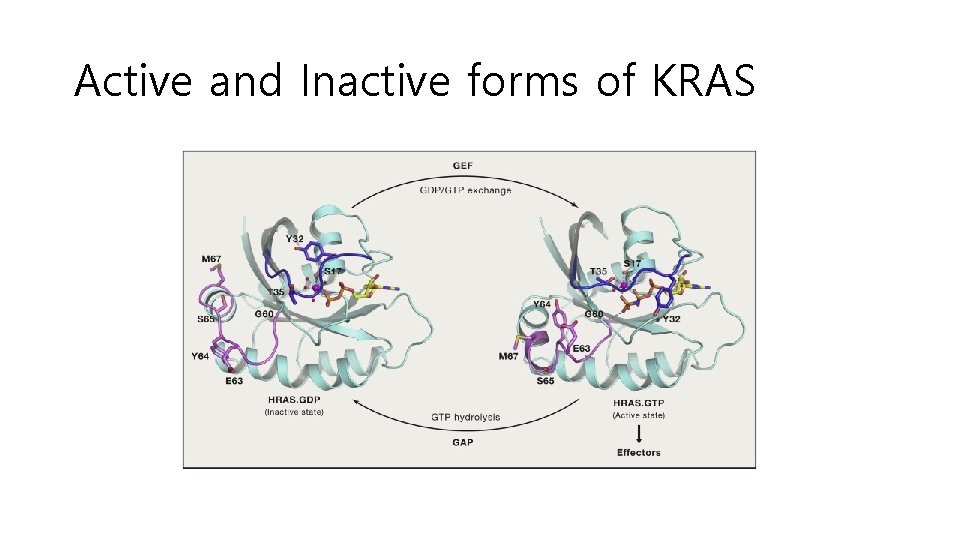
A mutated KRAS gene that is always "on" can promote cancer growth, despite treatment with drugs that target the upstream EGFR protein. Cancer-associated KRAS mutations (G 12 C, G 12 D, G 12 V, G 13 C, G 13 D, A 18 D, Q 61 H, K 117 N)

Active and Inactive forms of KRAS



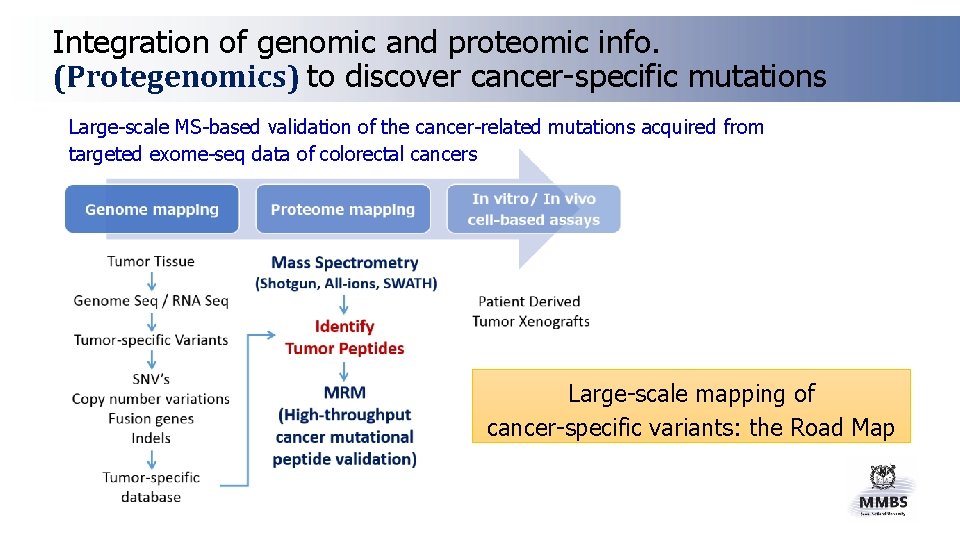
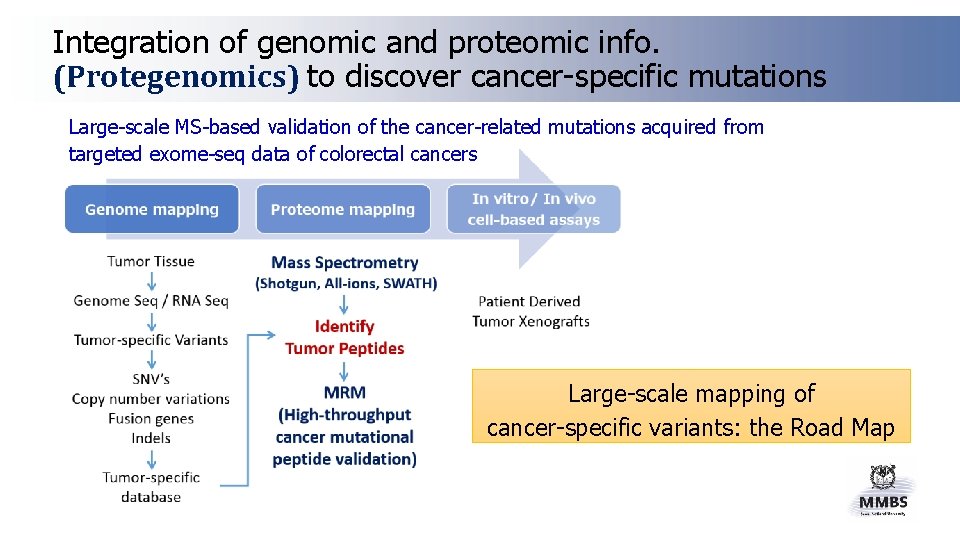
Integration of genomic and proteomic info. (Protegenomics) to discover cancer-specific mutations Large-scale MS-based validation of the cancer-related mutations acquired from targeted exome-seq data of colorectal cancers Large-scale mapping of cancer-specific variants: the Road Map

Cancer Moonshot initiative

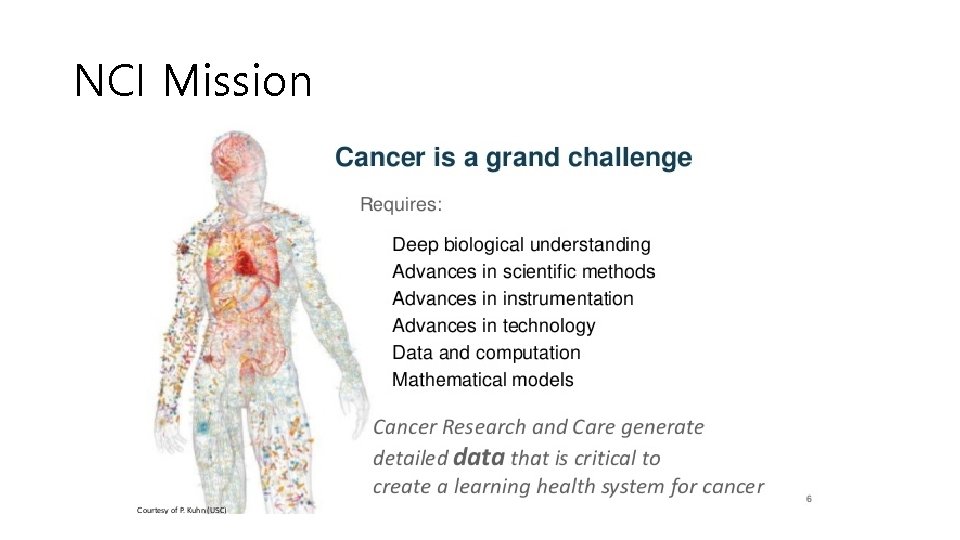
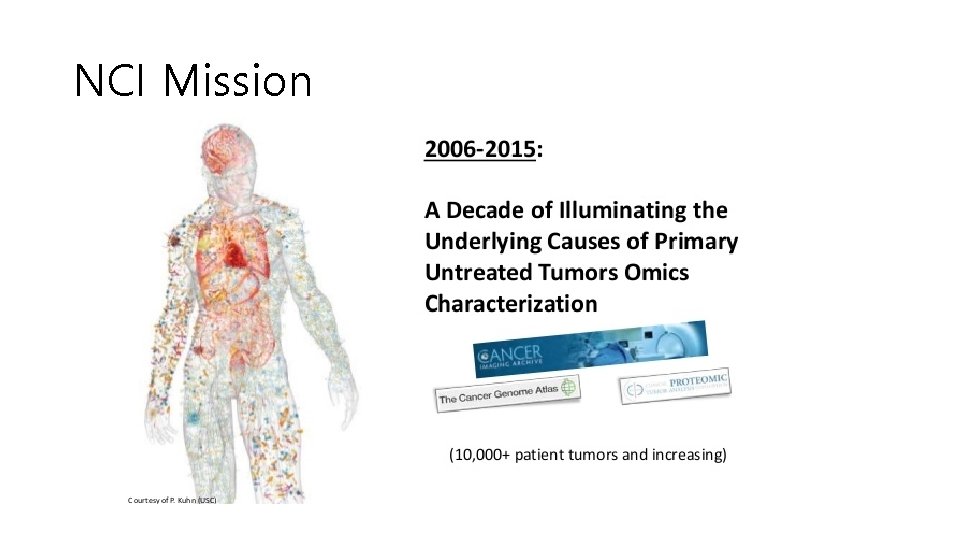
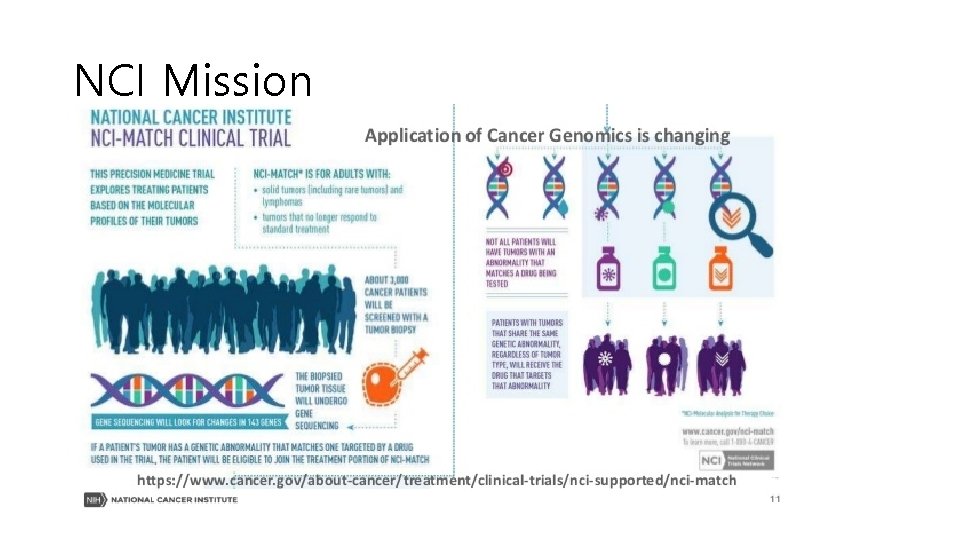
NCI (National Cancer Institute) Mission

NCI Mission

NCI Mission

NCI Mission

NCI Mission

NCI Mission

NCI Mission

NCI Mission

NCI Mission

NCI Mission

NCI Mission

NCI Mission

A single gene can give rise to multiple gene products ü RNA can be alternatively spliced or edited to form mature m. RNA ü Proteins are regulated by additional mechanisms such as posttranslational modifications, compartmentalization and proteolysis ü Biological function is determined by the complexity of these processes.

Proteomics § The abundance of an individual protein cannot be predicted in confidence by the level of the corresponding m. RNA § Initial studies in bacteria and yeast have suggested a reasonable correlation of about ~50 % between m. RNA and protein levels. In humans, global transcriptomic and proteomic analyses have shown that only 30 % of changes in protein levels can be explained by corresponding variations in m. RNA § This discrepancy emphasizes the importance of post-transcriptional regulations. Variable translation efficiency of m. RNA and protein levels, but the degradation and dynamic turnover of proteins are also involved. § Furthermore, the presence or absence of post-translational modifications, such as phosphorylation, glycosylation or ubiquitinylation, has a strong impact on protein stability, adding a further level of complexity § Together, the composition and dynamics of the proteome is not deducible from functional genomics data, and as a consequence proteomics is an indispensable and complementary approach to genomics and transcriptomics. Cellular and Molecular Life Sciences March 2015, Volume 72, Issue 5, pp 953– 957

Relationships between m. RNA and protein abundances, as observed in large-scale proteome- and transcriptome-profiling experiments l m. RNA transcript abundances only partially correlate with protein abundances (NIH 3 T 3 mouse fibroblast cells) l The correlation can be as little as 40% depending on the system. l Partially due to post-transcriptional and translational regulation, protein degradation, and RNA processing (alternative splicing and differential splicing) Nature Reviews Genetics 13, 227 -232

Course Description § Recent advances in genomic and proteomic technologies have led a huge increase in the generation of detail genomic and proteomic data more than ever. § There have been considerable efforts to disseminate and comprehend the data in the given biological conditions. § The objective of this course is to introduce students to the emerging field of integrated genomics and proteomics studies in biomedical research. § The topics to be discussed the current developments of genome sequencing and proteome technologies. The course will also emphasize a special interest on novel annotation fields, such as that of phenotypes, and highlights the recent efforts focused on the integrating gene annotations in the disease context.

Next Generation Sequencing § Rapid, inexpensive and high-throughput DNA sequencing capability § Post transcriptional regulation, including variable translation efficiency of m. RNA, regulatory actions as well as the dynamic turnover of proteins and their degradation, account for a general lack of correspondence between the variation of m. RNA levels and those of the corresponding protein

Next Generation Sequencing (NGS; Second Generation Sequencing and Third Generation Sequencing) Microarray 기반의 발현 연구에서 벗어나, RNASeq에 의한 전사체의 연구는 차등 발현 정보 이외 에 Fusion Gene, Novel Transcript/Gene, Alternative Splicing, RNA-DNA Differences(1) 등 의 추가적인 정보의 발굴이 가능 DNA 메틸화, 히스톤변형, mi. RNA와 같은 후성 유 전체의 경우에는 배아, 줄기세포, 암발생분화/줄기 세포를 중심으로 연구가 진행

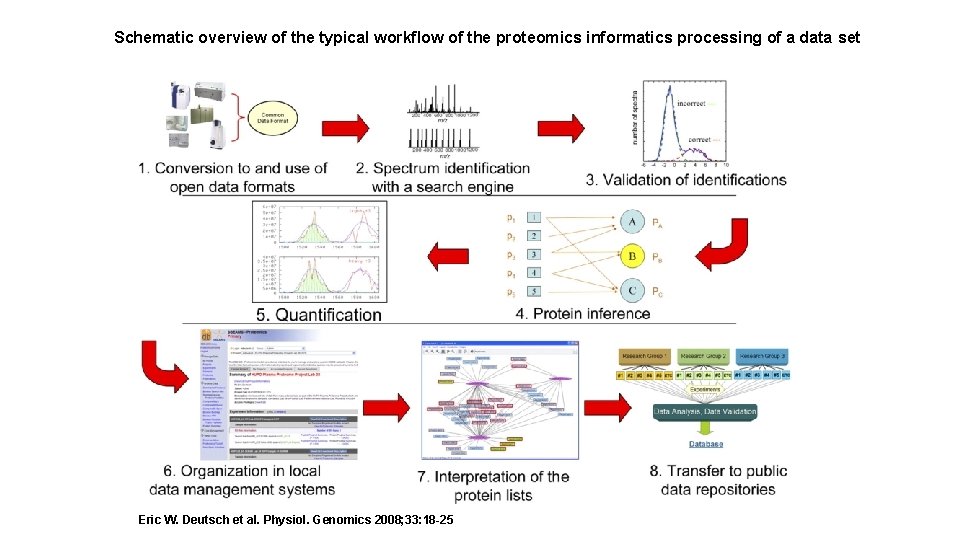
Schematic overview of the typical workflow of the proteomics informatics processing of a data set Eric W. Deutsch et al. Physiol. Genomics 2008; 33: 18 -25

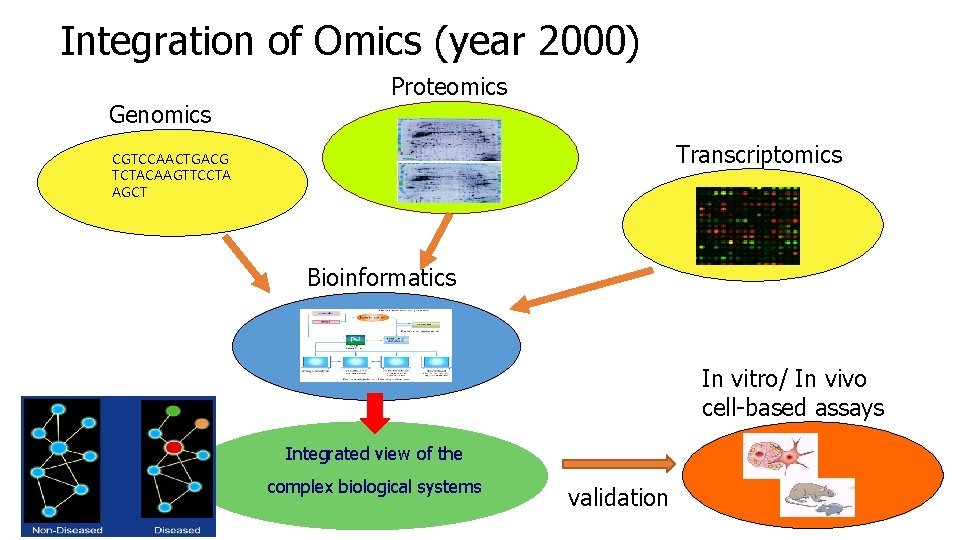
Integration of Omics (year 2000) Genomics Proteomics Transcriptomics CGTCCAACTGACG TCTACAAGTTCCTA AGCT Bioinformatics In vitro/ In vivo cell-based assays Integrated view of the complex biological systems validation

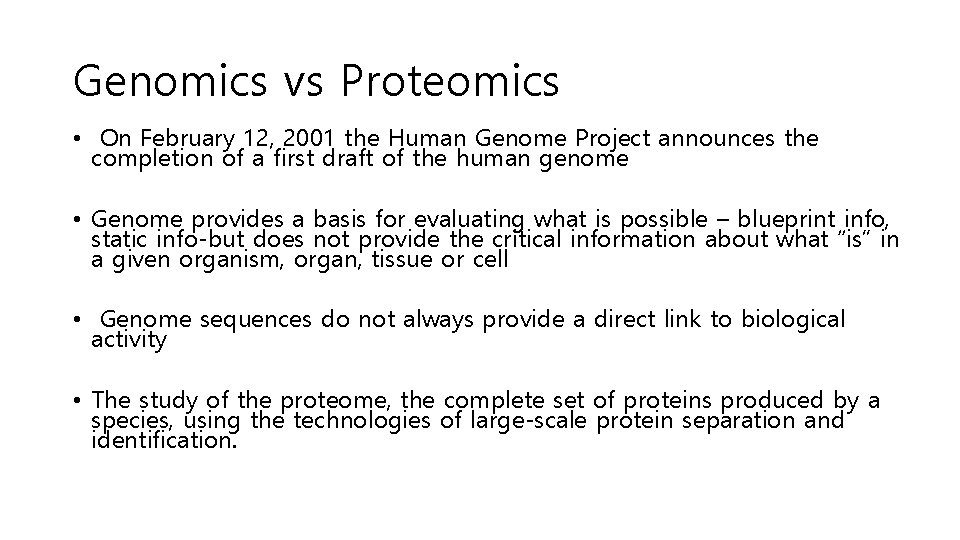
Genomics vs Proteomics • On February 12, 2001 the Human Genome Project announces the completion of a first draft of the human genome • Genome provides a basis for evaluating what is possible – blueprint info, static info-but does not provide the critical information about what “is” in a given organism, organ, tissue or cell • Genome sequences do not always provide a direct link to biological activity • The study of the proteome, the complete set of proteins produced by a species, using the technologies of large-scale protein separation and identification.

Concept of proteogenomics Genomic and transcriptomic data are used to generate customized protein sequence databases to help interpret proteomic data. In turn, the proteomic data provide protein-level validation of the gene expression data and help refine gene models. The enhanced gene models can help improve protein sequence databases for traditional proteomic analysis

Proteo. Genomics: Utilization of large-scale proteome data in genome annotation refinement and extension • Proteomics enable generation of accurate and comprehensive protein sequence of tissues, cells, and organisms • Proteomic approaches directly measure the peptides resulted from the expressed proteins and therefore, allow the confirmation /correction of the expressed coding regions in the genomic sequence Helmy et. al Genes to Cells 2012 • Thus, the proteome level information can be the desired source of information that complements the traditional genome analysis process

Proteo. Genomics: Utilization of large-scale proteome data in genome annotation refinement and extension • Possible to use proteogenomics to identify protein variants that could cause diseases, to identify protein biomarkers and to study genome variation • Recently, it became well acknowledged that the inclusion of the proteome data in the genome analysis, results in better genome annotation – ROUTINE PRACTICE Helmy et. al Genes to Cells 2012

Cancer moonshot

Proteomics data can be used to identify complete inventories of protein-coding genes (Proteogenomics)

Proteogenomic approach has become a complementary method for genome annotation

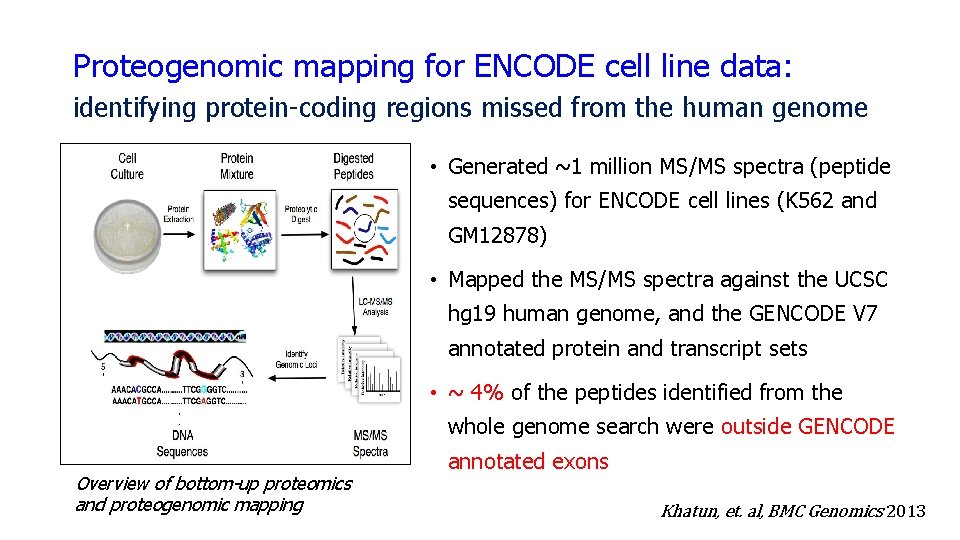
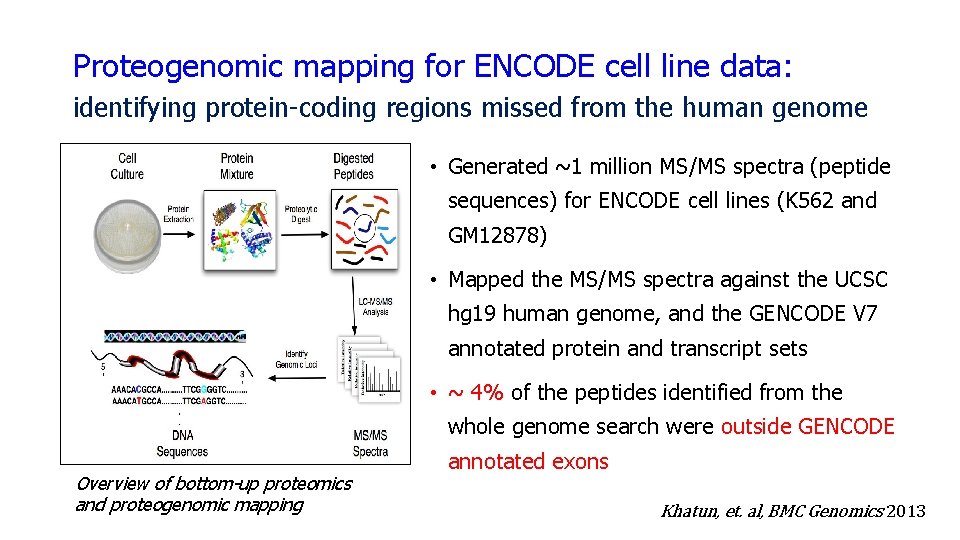
Proteogenomic mapping for ENCODE cell line data: identifying protein-coding regions missed from the human genome • Generated ~1 million MS/MS spectra (peptide sequences) for ENCODE cell lines (K 562 and GM 12878) • Mapped the MS/MS spectra against the UCSC hg 19 human genome, and the GENCODE V 7 annotated protein and transcript sets • ~ 4% of the peptides identified from the whole genome search were outside GENCODE Overview of bottom-up proteomics and proteogenomic mapping annotated exons Khatun, et. al, BMC Genomics 2013

Cancer Proteogenomics Cancer is characterized by altered expression of tumor drivers and suppressors • Results from gene mutations causing changes in protein expression, activity Cancer Proteomics • • Are genomic variants evident at the protein level? What is their effect on protein function associated cancers? What proteins have altered expression in tumor tissue? Can these proteins be used as cancer biomarkers for disease prevention, prognosis and targeted therapy?




Integration of genomic and proteomic info. (Protegenomics) to discover cancer-specific mutations Large-scale MS-based validation of the cancer-related mutations acquired from targeted exome-seq data of colorectal cancers Large-scale mapping of cancer-specific variants: the Road Map

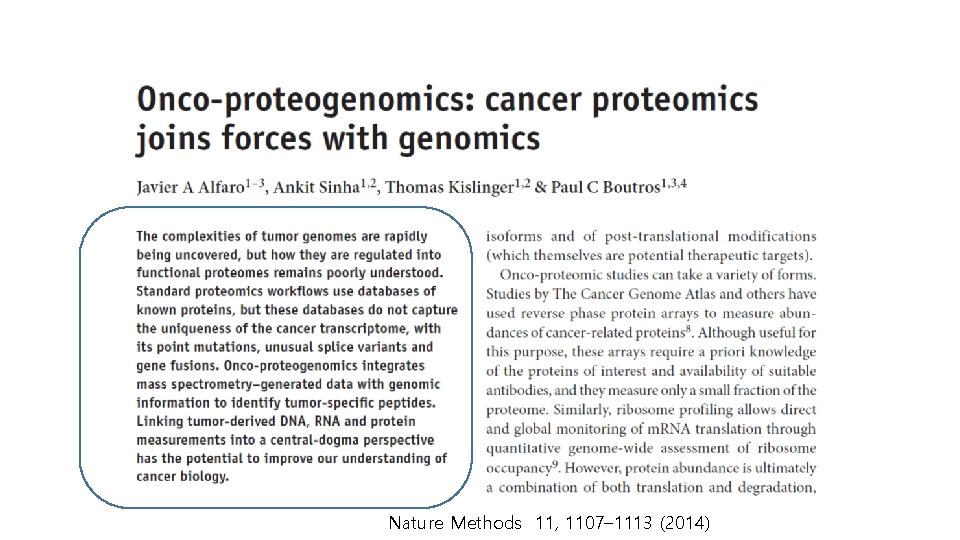
Nature Methods 11, 1107– 1113 (2014)

Proteogenomics 최근논문 Proteogenomics: Integrating Next-Generation Sequencing and Mass Spectrometry to Characterize Human Proteomic Variation-Annu Rev Anal Chem (Palo Alto Calif). Author manuscript; available in PMC 2016 August 19 Proteogenomic convergence for understanding cancer pathways and networks -Clinical Proteomics 2014 Methods, tools and current perspectives in proteogenomics –MCP 2017 Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer-cell 2016 First Large-Scale Proteogenomic Study of Breast Cancer Provides Insight into Potential Therapeutic Targets-Nature 2016 Proteogenomic characterization of human colon and rectal cancer- Nature 2014

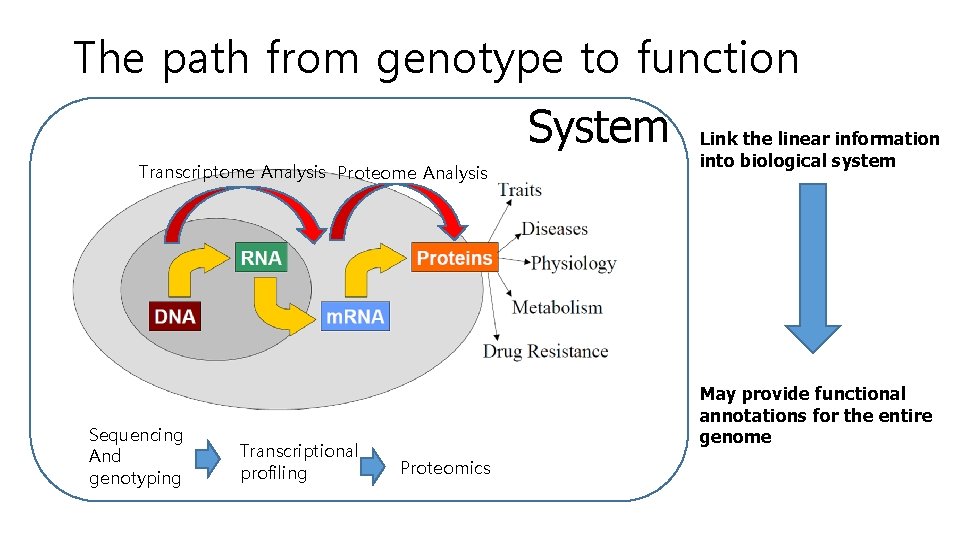
The path from genotype to function System Transcriptome Analysis Proteome Analysis Sequencing And genotyping Transcriptional profiling Link the linear information into biological system May provide functional annotations for the entire genome Proteomics

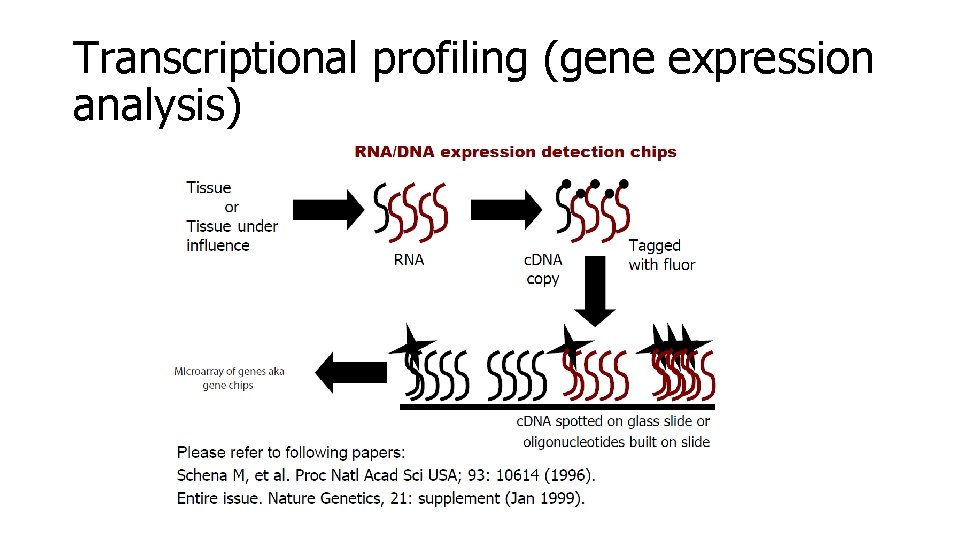
Transcriptional profiling (gene expression analysis)

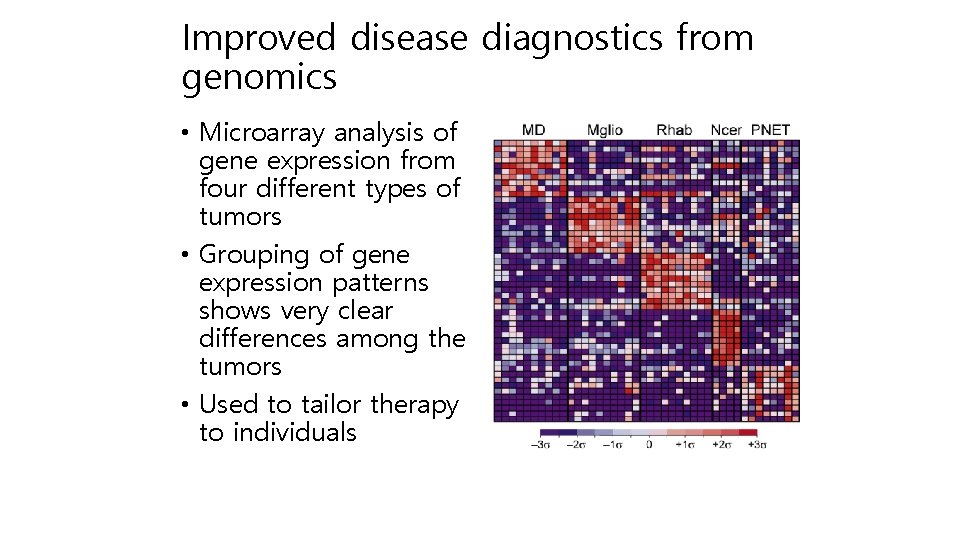
Improved disease diagnostics from genomics • Microarray analysis of gene expression from four different types of tumors • Grouping of gene expression patterns shows very clear differences among the tumors • Used to tailor therapy to individuals

Prescreening based on genomes All patients with same diagnosis 1 Remove Toxic and Nonresponders 2 Treat Responders and Patients Not Predisposed to Toxic

Relationships between m. RNA and protein abundances, as observed in large-scale proteome- and transcriptome-profiling experiments l m. RNA transcript abundances only partially correlate with protein abundances (NIH 3 T 3 mouse fibroblast cells) l The correlation can be as little as 40% depending on the system. l Partially due to post-transcriptional and translational regulation, protein degradation, and RNA processing (alternative splicing and differential splicing) Nature Reviews Genetics 13, 227 -232

Proteomics - Type of Studies Which Proteins, when, where, with whom? • Protein Mining – Catalog proteins, genome annotation and gene function prediction in a tissue or cell etc. • Differential Protein Expression Profiling – Identification of proteins in a sample as a function of a particular state: differentiation, stage of development, disease state, response to drug or stimulus • Subcellular location / Secretion • Mapping protein modifications – Characterization of PTM (phosphorylation, glycosylation, ubiquination. oxidation etc) • Protein Interactions – Ligand-Receptor & Oligomerization and protein-DNA • Network-based analysis – Identification of proteins in functional networks: biosynthetic pathways, signal transduction pathways, multiprotein complexes

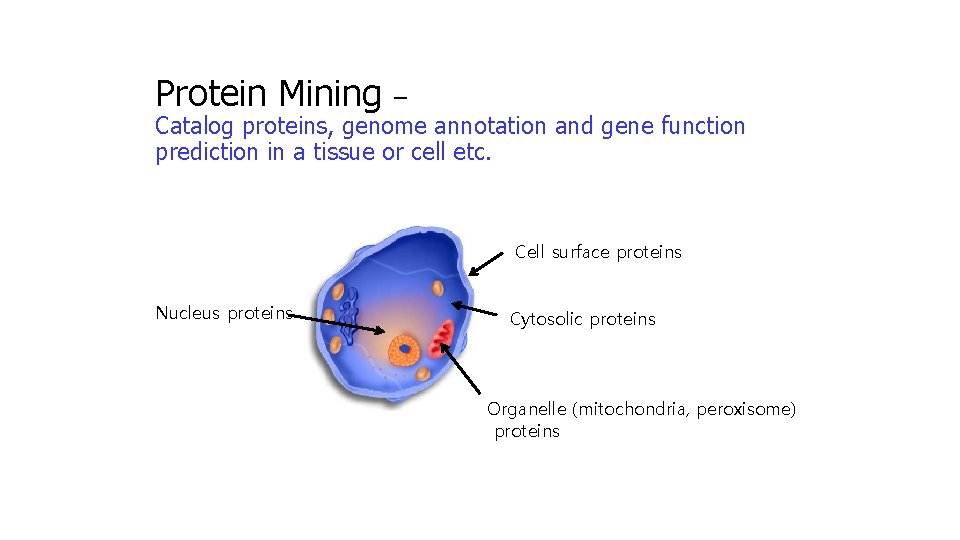
Protein Mining – Catalog proteins, genome annotation and gene function prediction in a tissue or cell etc. Cell surface proteins Nucleus proteins Cytosolic proteins Organelle (mitochondria, peroxisome) proteins

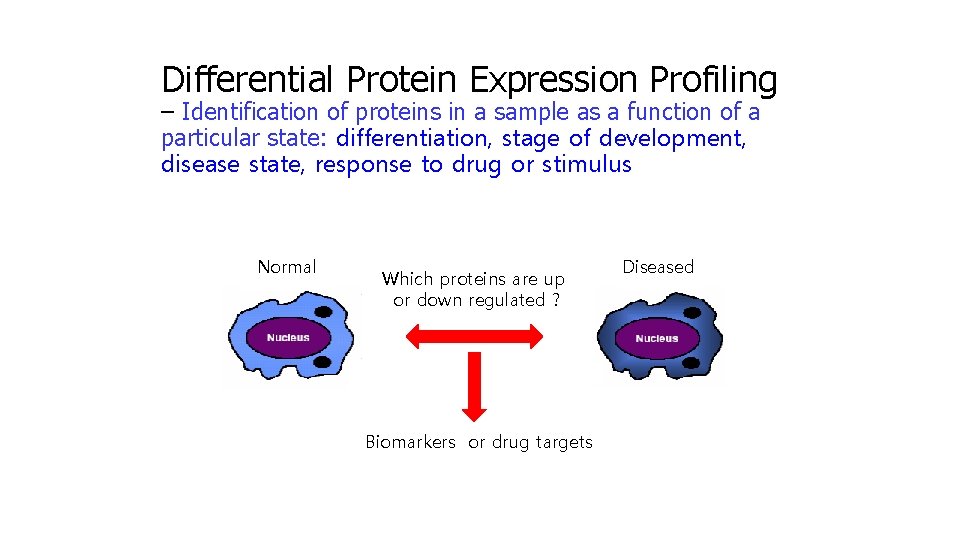
Differential Protein Expression Profiling – Identification of proteins in a sample as a function of a particular state: differentiation, stage of development, disease state, response to drug or stimulus Normal Which proteins are up or down regulated ? Biomarkers or drug targets Diseased

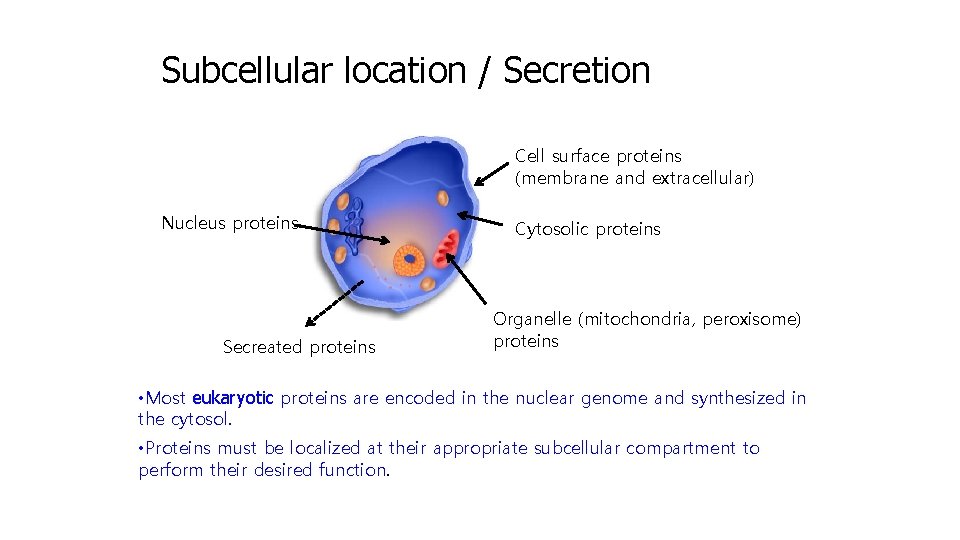
Subcellular location / Secretion Cell surface proteins (membrane and extracellular) Nucleus proteins Secreated proteins Cytosolic proteins Organelle (mitochondria, peroxisome) proteins • Most eukaryotic proteins are encoded in the nuclear genome and synthesized in the cytosol. • Proteins must be localized at their appropriate subcellular compartment to perform their desired function.

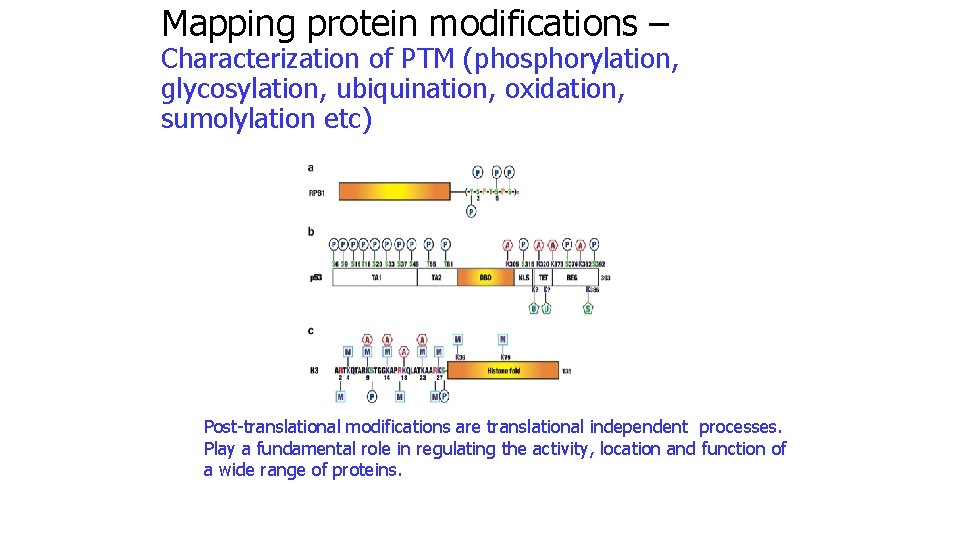
Mapping protein modifications – Characterization of PTM (phosphorylation, glycosylation, ubiquination, oxidation, sumolylation etc) Post-translational modifications are translational independent processes. Play a fundamental role in regulating the activity, location and function of a wide range of proteins.

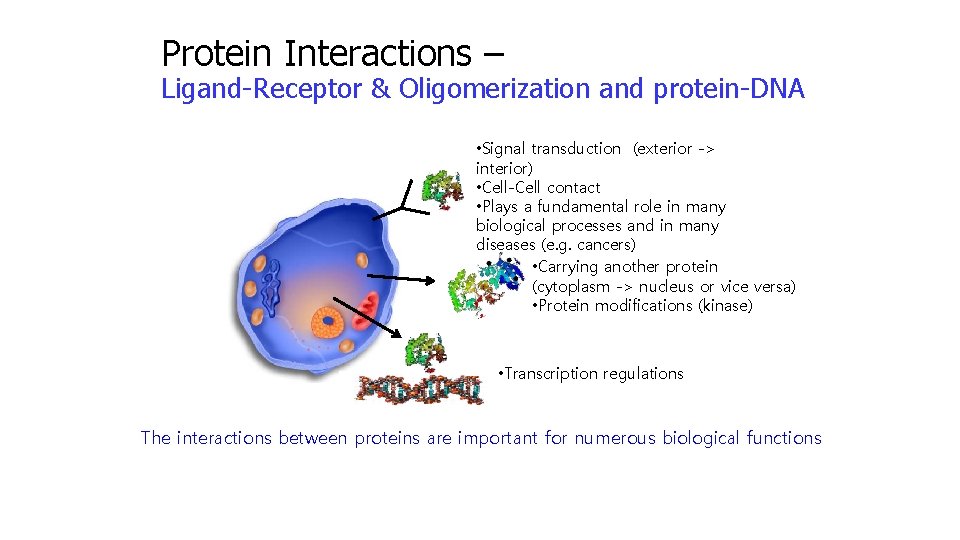
Protein Interactions – Ligand-Receptor & Oligomerization and protein-DNA • Signal transduction (exterior -> interior) • Cell-Cell contact • Plays a fundamental role in many biological processes and in many diseases (e. g. cancers) • Carrying another protein (cytoplasm -> nucleus or vice versa) • Protein modifications (kinase) • Transcription regulations The interactions between proteins are important for numerous biological functions

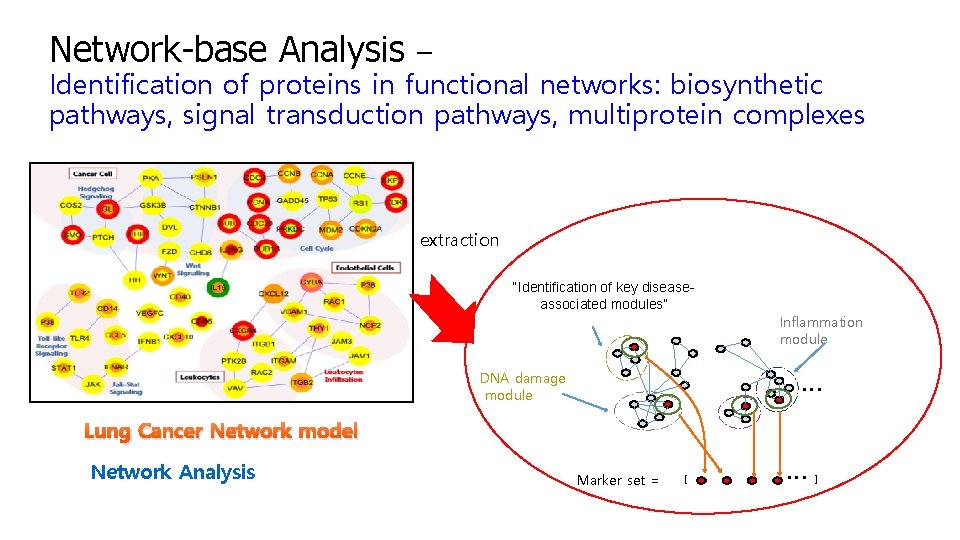
Network-base Analysis – Identification of proteins in functional networks: biosynthetic pathways, signal transduction pathways, multiprotein complexes extraction “Identification of key diseaseassociated modules” Inflammation module … DNA damage module Lung Cancer Network model Network Analysis Marker set = [ … ]

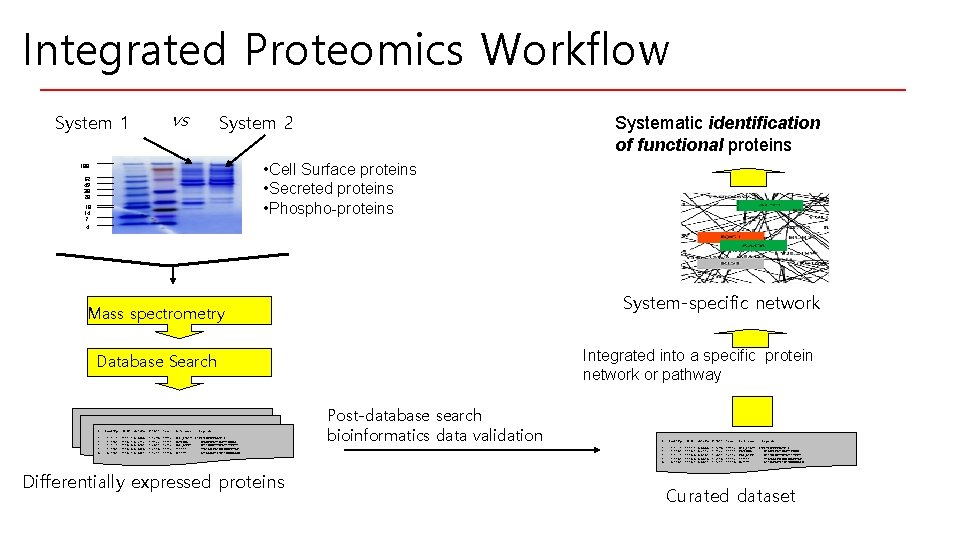
Integrated Proteomics Workflow vs System 1 System 2 Systematic identification of functional proteins • Cell Surface proteins • Secreted proteins • Phospho-proteins 188 62 49 38 28 18 14 7 4 System-specific network Mass spectrometry Integrated into a specific protein network or pathway Database Search # --1. 2. 3. 4. 5. Rank/Sp ------1 2 3 4 5 / 1 /403 / 3 /209 /381 (M+H)+ delt. Cn ------1994. 3 0. 0000 1995. 1 0. 6126 1995. 0 0. 5952 1995. 0 0. 5895 1995. 1 0. 5514 C*10^4 ------ Ions ---- Reference ----- Peptide ------- 4. 4675 2. 7366 2. 6591 2. 6335 2. 4634 17/26 13/34 16/36 14/36 13/38 G 3 P_RABIT (R)VPTPNVSVVDLTC*R SLTRNGL (E)LGKPVLTANQVTIWEGLR FLP_LACCA (N)IANPNVYTETLTAATVCTI A 42912 (Y)LALLPSDAEGPHGQFVTDK H 69373 (L)ALLVLVAPAMAAGNGEDLRN Differentially expressed proteins Post-database search bioinformatics data validation # --1. 2. 3. 4. 5. Rank/Sp ------1 / 1 2 /403 3 / 3 4 /209 5 /381 (M+H)+ -----1994. 3 1995. 1 1995. 0 1995. 1 delt. Cn -----0. 0000 0. 6126 0. 5952 0. 5895 0. 5514 C*10^4 -----4. 4675 2. 7366 2. 6591 2. 6335 2. 4634 Ions ---17/26 13/34 16/36 14/36 13/38 Reference Peptide --------G 3 P_RABIT (R)VPTPNVSVVDLTC*R SLTRNGL (E)LGKPVLTANQVTIWEGLR FLP_LACCA (N)IANPNVYTETLTAATVCTI A 42912 (Y)LALLPSDAEGPHGQFVTDK H 69373 (L)ALLVLVAPAMAAGNGEDLRN Curated dataset

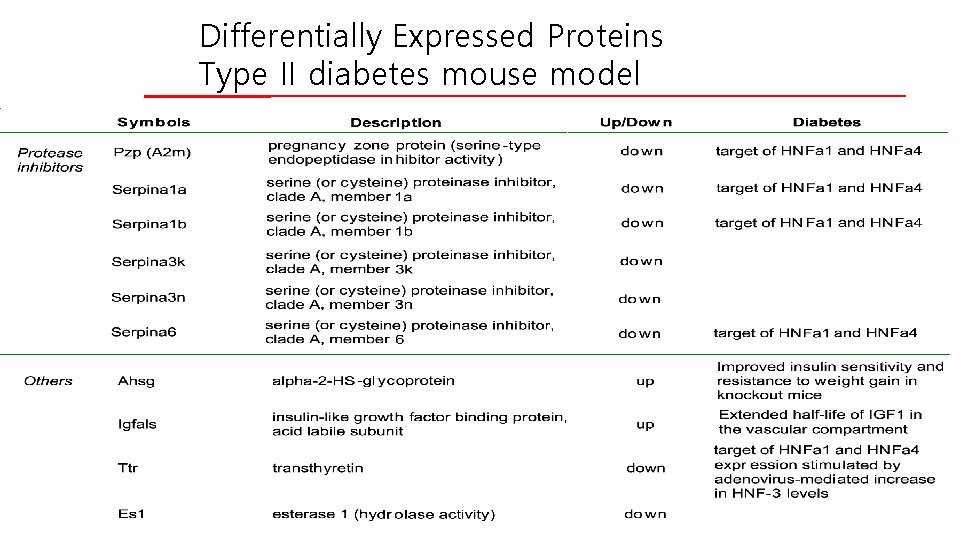
Differentially Expressed Proteins Type II diabetes mouse model

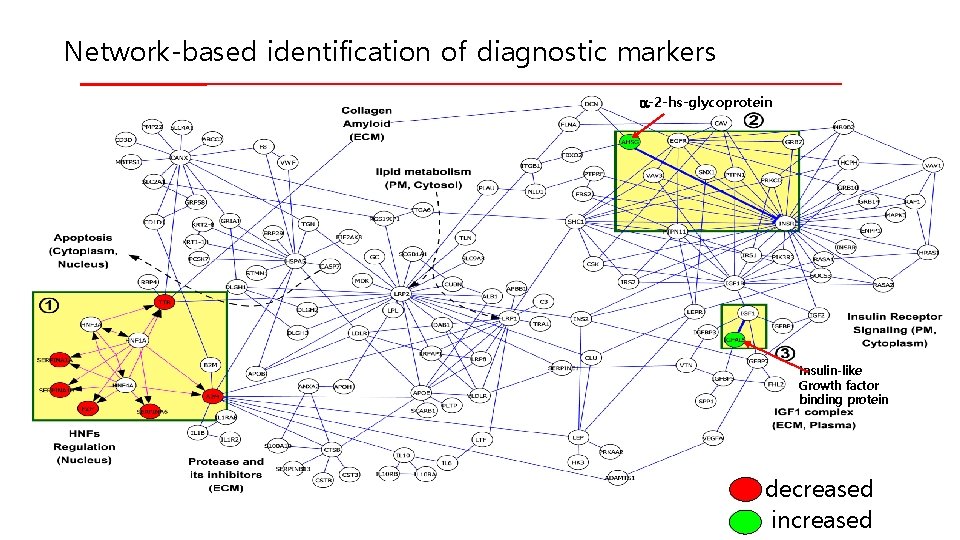
Network-based identification of diagnostic markers a-2 -hs-glycoprotein Insulin-like Growth factor binding protein decreased increased

Next Generation Sequencing § Rapid, inexpensive and high-throughput DNA sequencing capability offers an opportunity to sequence whole genomes of a large number of species (http: //bio. lmu. de/~parsch/evogen/Next. Ge n. pdf) § However, the cataloging of proteincoding genes from these species remains a non-trivial task with the majority of initial genome annotation dependent on the use of gene prediction algorithms

Next Generation Sequencing (NGS; Second Generation Sequencing and Third Generation Sequencing) Microarray 기반의 발현 연구에서 벗어나, RNASeq에 의한 전사체의 연구는 차등 발현 정보 이외 에 Fusion Gene, Novel Transcript/Gene, Alternative Splicing, RNA-DNA Differences(1) 등 의 추가적인 정보의 발굴이 가능 DNA 메틸화, 히스톤변형, mi. RNA와 같은 후성 유 전체의 경우에는 배아, 줄기세포, 암발생분화/줄기 세포를 중심으로 연구가 진행

Proteo. Genomics: Utilization of large-scale proteome data in genome annotation refinement and extension • Proteomics enable generation of accurate and comprehensive protein sequence of tissues, cells, and organisms • Proteomic approaches directly measure the peptides resulted from the expressed proteins and therefore, allow the confirmation /correction of the expressed coding regions in the genomic sequence Helmy et. al Genes to Cells 2012 • Thus, the proteome level information can be the desired source of information that complements the traditional genome analysis process

Proteo. Genomics: Utilization of large-scale proteome data in genome annotation refinement and extension • Possible to use proteogenomics to identify protein variants that could cause diseases, to identify protein biomarkers and to study genome variation • Recently, it became well acknowledged that the inclusion of the proteome data in the genome analysis, results in better genome annotation – ROUTINE PRACTICE Helmy et. al Genes to Cells 2012

Proteomics data can be used to identify complete inventories of protein-coding genes (Proteogenomics)

Proteogenomic approach has become a complementary method for genome annotation

Proteogenomic mapping for ENCODE cell line data: identifying protein-coding regions missed from the human genome • Generated ~1 million MS/MS spectra (peptide sequences) for ENCODE cell lines (K 562 and GM 12878) • Mapped the MS/MS spectra against the UCSC hg 19 human genome, and the GENCODE V 7 annotated protein and transcript sets • ~ 4% of the peptides identified from the whole genome search were outside GENCODE Overview of bottom-up proteomics and proteogenomic mapping annotated exons Khatun, et. al, BMC Genomics 2013