Topic 2 Molecular Biology 2 7 DNA replication

Topic 2 Molecular Biology

2. 7 DNA replication, transcription & translation
Bellwork 1. With your partner, discuss the differences between DNA and RNA. 2. Then place the following steps of transcription in the correct order: a. After the gene has completely transcribed, the m. RNA breaks away from the DNA and leaves the nucleus for the ribosomes b. The DNA double helix reforms c. The polynucleotide DNA strand with the gene of interest acts as a template for m. RNA. d. RNA polymerase adds free RNA nucleotide base pairs to DNA nucleotides, forming phosphodiester bonds between RNA nucleotides. e. The DNA helix is opened at the position of the gene by RNA polymerase

Steps of Transcription 1. The DNA helix is opened at the position of the gene by RNA polymerase 2. The polynucleotide DNA strand with the gene of interest acts as a template for m. RNA. 3. RNA polymerase adds free RNA nucleotide base pairs to DNA nucleotides, forming phosphodiester bonds between RNA nucleotides. 4. After the gene has completely transcribed, the m. RNA breaks away from the DNA and leaves the nucleus for the ribosomes. 5. The DNA double helix reforms.

2. 7 DNA Replication, Transcription, Translation Give purpose and product of the following: • Replication: • Transcription: • Translation:
REPLICATION: 1 DNA to 2 DNA
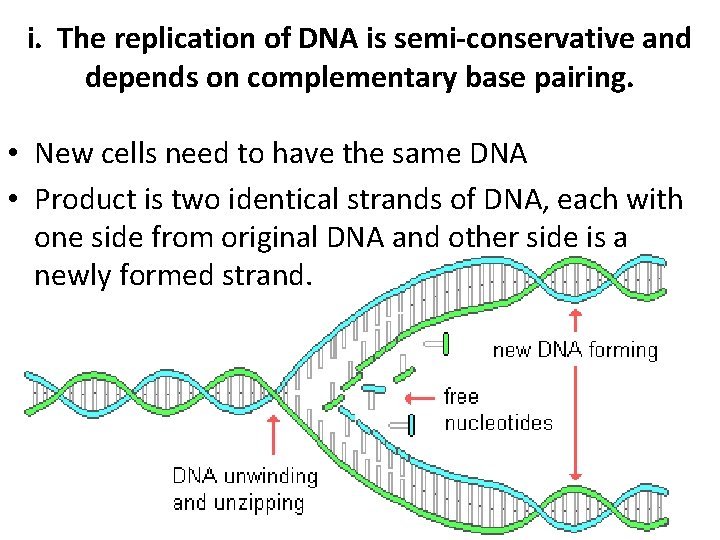
i. The replication of DNA is semi-conservative and depends on complementary base pairing. • New cells need to have the same DNA • Product is two identical strands of DNA, each with one side from original DNA and other side is a newly formed strand.
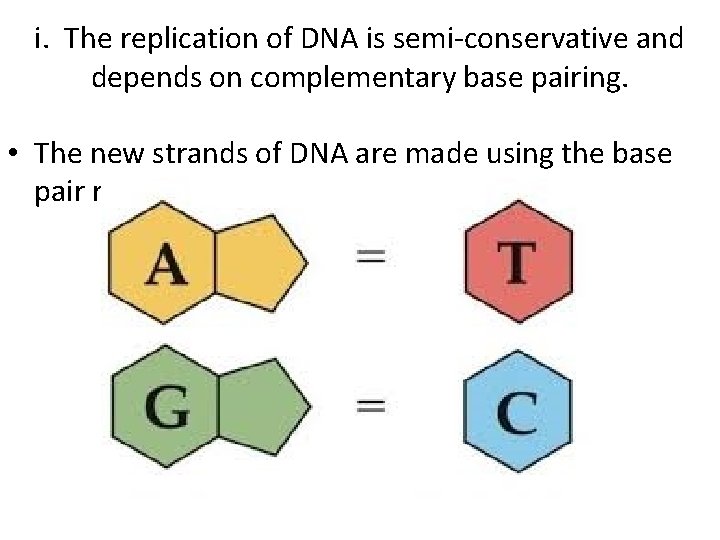
i. The replication of DNA is semi-conservative and depends on complementary base pairing. • The new strands of DNA are made using the base pair rules.
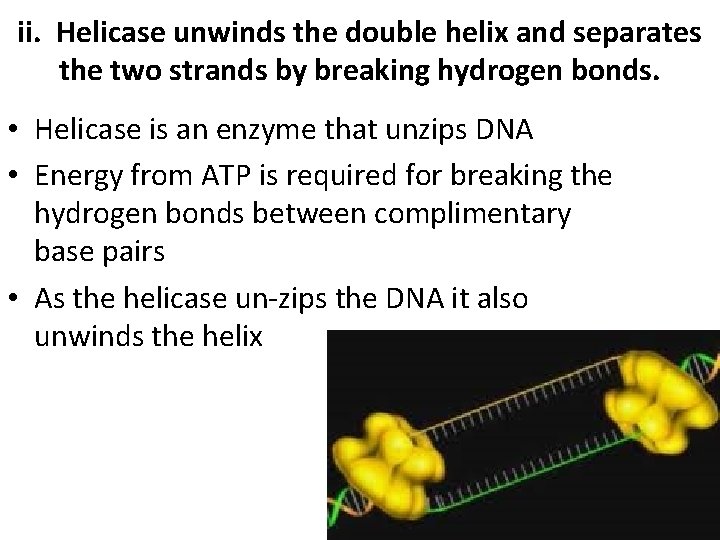
ii. Helicase unwinds the double helix and separates the two strands by breaking hydrogen bonds. • Helicase is an enzyme that unzips DNA • Energy from ATP is required for breaking the hydrogen bonds between complimentary base pairs • As the helicase un-zips the DNA it also unwinds the helix
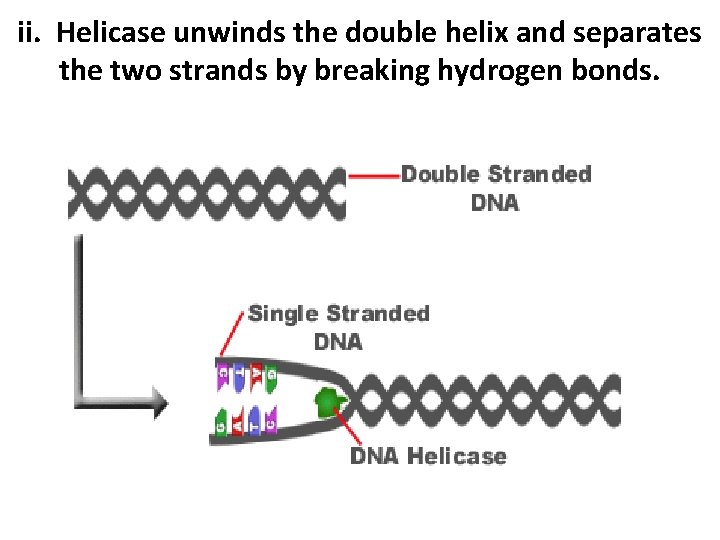
ii. Helicase unwinds the double helix and separates the two strands by breaking hydrogen bonds.
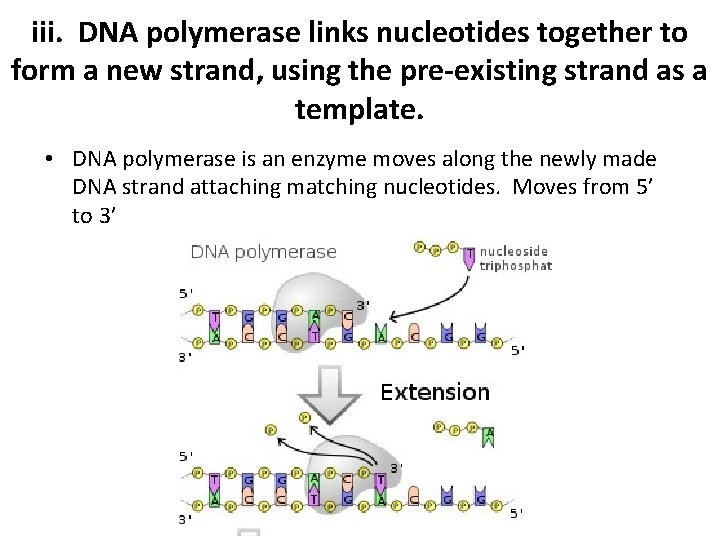
iii. DNA polymerase links nucleotides together to form a new strand, using the pre-existing strand as a template. • DNA polymerase is an enzyme moves along the newly made DNA strand attaching matching nucleotides. Moves from 5’ to 3’
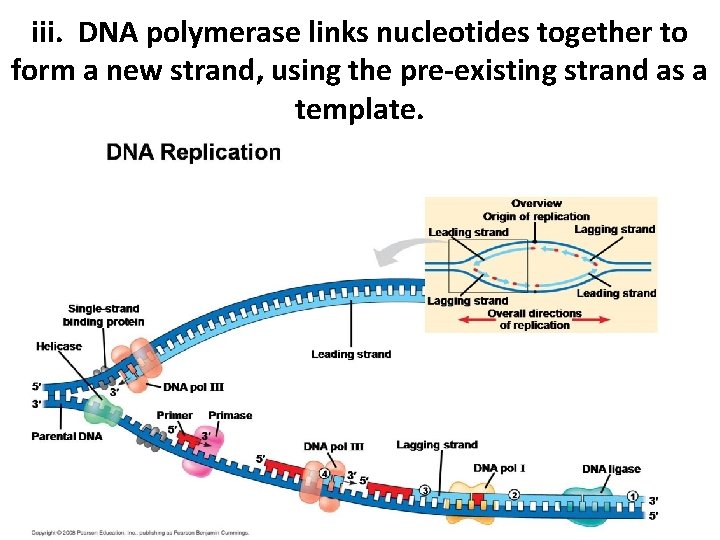
iii. DNA polymerase links nucleotides together to form a new strand, using the pre-existing strand as a template.
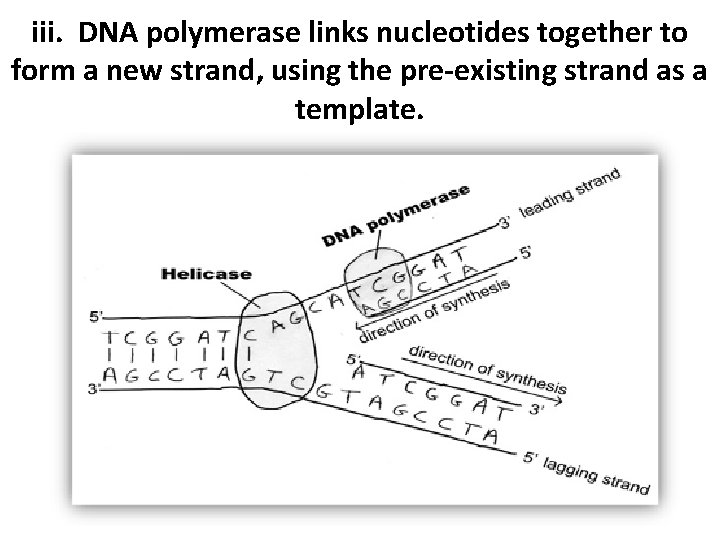
iii. DNA polymerase links nucleotides together to form a new strand, using the pre-existing strand as a template.

Replication in ACTION DNA Replication 3 D 5. 45 min https: //www. youtube. com/watch? v=27 Tx. Ko. FU 2 Nw DNA structure and replication: Crash course 12: 58 https: //www. youtube. com/watch? v=8 k. K 2 zwj. RV 0 M http: //www. youtube. com/watch? v=yq. ESR 7 E 4 b_8
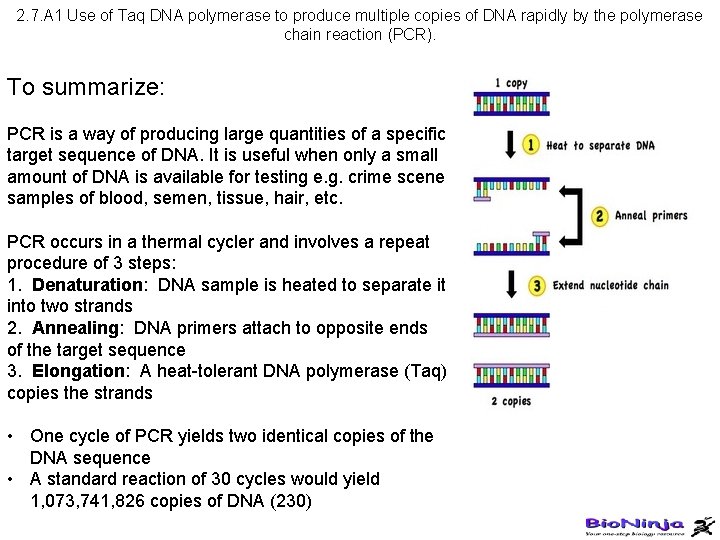
2. 7. A 1 Use of Taq DNA polymerase to produce multiple copies of DNA rapidly by the polymerase chain reaction (PCR). To summarize: PCR is a way of producing large quantities of a specific target sequence of DNA. It is useful when only a small amount of DNA is available for testing e. g. crime scene samples of blood, semen, tissue, hair, etc. PCR occurs in a thermal cycler and involves a repeat procedure of 3 steps: 1. Denaturation: DNA sample is heated to separate it into two strands 2. Annealing: DNA primers attach to opposite ends of the target sequence 3. Elongation: A heat-tolerant DNA polymerase (Taq) copies the strands • One cycle of PCR yields two identical copies of the DNA sequence • A standard reaction of 30 cycles would yield 1, 073, 741, 826 copies of DNA (230)
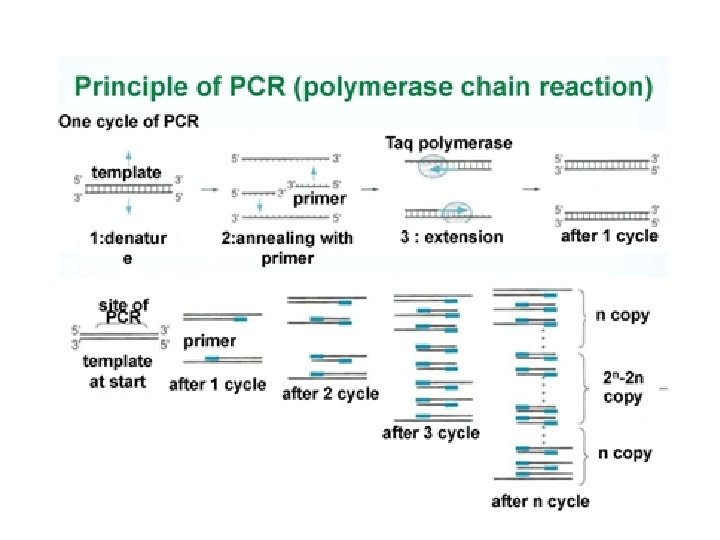
2. 7. A 1 Use of Taq DNA polymerase to produce multiple copies of DNA rapidly by the polymerase chain reaction (PCR). After clicking on the my. DNA link choose techniques and then amplifying to access the tutorials on the polymerase chain reaction (PCR). Alternatively watch the Mc. Graw-Hill tutorial http: //www. dnai. org/b/index. html http: //highered. mcgraw-hill. com/olc/dl/120078/micro 15. swf
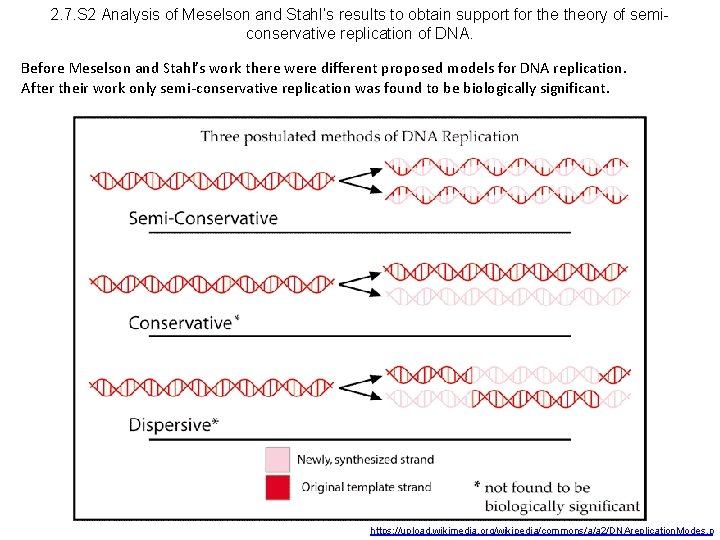
2. 7. S 2 Analysis of Meselson and Stahl’s results to obtain support for theory of semiconservative replication of DNA. Before Meselson and Stahl’s work there were different proposed models for DNA replication. After their work only semi-conservative replication was found to be biologically significant. https: //upload. wikimedia. org/wikipedia/commons/a/a 2/DNAreplication. Modes. p

SKILL: Analysis of Meselson and Stahl’s results to obtain support for theory of semi-conservative replication of DNA> • Read through the article for Obtaining evidence for theory of semi-conservative replication and complete the data based question on the Analysis of Meselson and Stahl’s results on page 113 - and 114
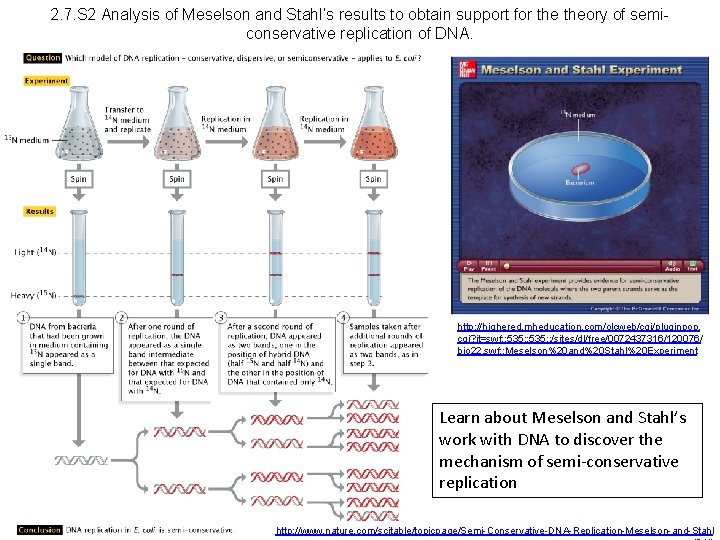
2. 7. S 2 Analysis of Meselson and Stahl’s results to obtain support for theory of semiconservative replication of DNA. http: //highered. mheducation. com/olcweb/cgi/pluginpop. cgi? it=swf: : 535: : /sites/dl/free/0072437316/120076/ bio 22. swf: : Meselson%20 and%20 Stahl%20 Experiment Learn about Meselson and Stahl’s work with DNA to discover the mechanism of semi-conservative replication http: //www. nature. com/scitable/topicpage/Semi-Conservative-DNA-Replication-Meselson-and-Stahl
2. 7. S 2 Analysis of Meselson and Stahl’s results to obtain support for theory of semiconservative replication of DNA. At the start of a Meselson and Stahl experiment (generation 0) a single band of DNA with a density of 1. 730 g cm-3 was found. After 4 generations two bands were found, but the main band had a density of 1. 700 g cm-3. a. Explain why the density of the main band changed over four generations. (2) b. After one generation one still only one DNA band appears, but the density has changed. i. Estimate the density of the band. (1) ii. Which (if any) mechanisms of DNA replication are falsified by this result? (1) iii. Explain why the identified mechanism(s) are falsified. (1) c. Describe the results after two generations and which mechanisms and explain the identified mechanism(s) (if any) are falsified as a consequence. (3) d. Describe and explain the result found by centrifuging a mixture of DNA from generation 0 and 2. (2)

2. 7. S 2 Analysis of Meselson and Stahl’s results to obtain support for theory of semiconservative replication of DNA. At the start of a Meselson and Stahl experiment (generation 0) a single band of DNA with a density of 1. 730 g cm-3 was found. After 4 generations two bands were found, but the main band had a density of 1. 700 g cm-3. a. Explain why the density of the main band changed over four generations. (2) • N 15 isotope has a greater mass than N 14 isotope due to the extra neutron • Generation 0 contained DNA with exclusively N 15 isotopes (giving it a greater density) • With each generation the proportion N 14 isotope (from free nucleotides) increases as the mass of DNA doubles • After four generations most strands contain only N 14 isotope – the dominant band at a density of 1. 700 g cm-3. • N 15 isotope remains, but is combined in strands with N 14 isotope – a second band at a density between 1. 730 and 1. 700 g cm-3.

2. 7. S 2 Analysis of Meselson and Stahl’s results to obtain support for theory of semiconservative replication of DNA. At the start of a Meselson and Stahl experiment (generation 0) a single band of DNA with a density of 1. 730 g cm-3 was found. After 4 generations two bands were found, but the main band had a density of 1. 700 g cm-3. b. After one generation one still only one DNA band appears, but the density has changed. i. Estimate the density of the band. (1) • The band would contain equally amounts of N 14 isotope and N 15 isotope • Density of an all N 15 isotope band is 1. 730 g cm-3. • Density of an all N 14 isotope band is 1. 700 g cm-3. • Density of an the mixed isotope band is the average of the two: = ( 1. 730 g cm-3 + 1. 700 g cm-3 ) / 2 = 1. 715 g cm-3 ii. Which (if any) mechanisms of DNA replication are falsified by this result? (1) • conservative replication iii. Explain why the identified mechanism(s) are falsified. (1) • For conservative replication to be the case two bands should appear in all generations after generation 0
2. 7. S 2 Analysis of Meselson and Stahl’s results to obtain support for theory of semiconservative replication of DNA. At the start of a Meselson and Stahl experiment (generation 0) a single band of DNA with a density of 1. 730 g cm-3 was found. After 4 generations two bands were found, but the main band had a density of 1. 700 g cm-3. c. Describe the results after two generations and which mechanisms and explain the identified mechanism(s) (if any) are falsified as a consequence. (3) • 2 bands: • One band containing a mixture of N 15 and N 14 isotopes – semiconservative replication preserves the DNA strands containing N 15 isotopes, but combines them with N 14 nucleotides during replication. • One band containing all N 14 isotopes - during replication from generation 1 to generation 2. The new strands consisting of of N 14 isotopes are replicated using N 14 nucleotides creating strands containing just N 14 isotopes. • Dispersive replication is falsified as this model would continue to produce a single band, containing proportionally less N 15 isotope.
2. 7. S 2 Analysis of Meselson and Stahl’s results to obtain support for theory of semiconservative replication of DNA. At the start of a Meselson and Stahl experiment (generation 0) a single band of DNA with a density of 1. 730 g cm-3 was found. After 4 generations two bands were found, but the main band had a density of 1. 700 g cm-3. d. Describe and explain the result found by centrifuging a mixture of DNA from generation 0 and 2. (2) • 3 bands: • One band from generation 0 containing all N 15 isotopes – no replication has occured • One band from generation 2 containing a mixture of N 15 and N 14 isotopes – semi-conservative replication preserves the DNA strands containing N 15 isotopes, but combines them with N 14 nucleotides during replication. • One band from generation 2 (all replicated DNA) containing all N 14 isotopes - during replication from generation 1 to generation 2. The new strands consisting of of N 14 isotopes are replicated using N 14 nucleotides creating strands containing just N 14 isotopes.
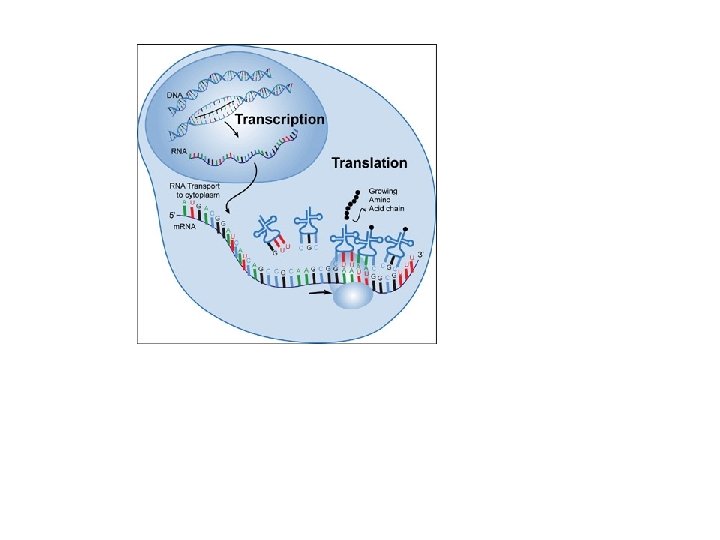
Q - What is the purpose of transcription and translation? AThese processes work together to create a polypeptide which in turns folds to become a protein. Proteins carry many essential functions in cells. For more detail review 2. 4. U 7 Living organisms synthesize many different proteins with a wide range of functions. Catalysis Tensile strengthening Muscle contraction Transport of nutrients and gases Use the learn. genetics tutorial to discover one example: Cell adhesion Hormones Receptors Cytoskeletons Packing of DNA Blood clotting Immunity Membrane transport http: //learn. genetics. utah. edu/content/molecules/firefly/
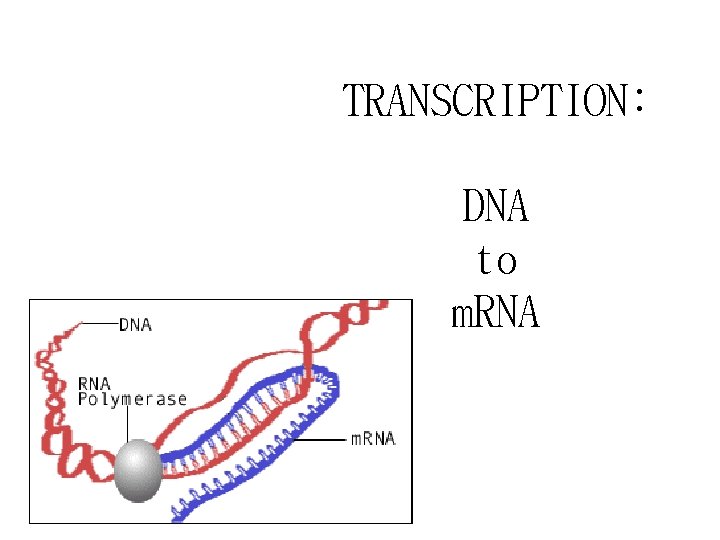
TRANSCRIPTION: DNA to m. RNA
iv. Transcription is the synthesis of m. RNA copied from the DNA base sequence by RNA polymerase • Genes are the instructions for making proteins • Transcription is the synthesis of RNA using DNA as a template • RNA is single-stranded so transcription only occurs along one strand of DNA
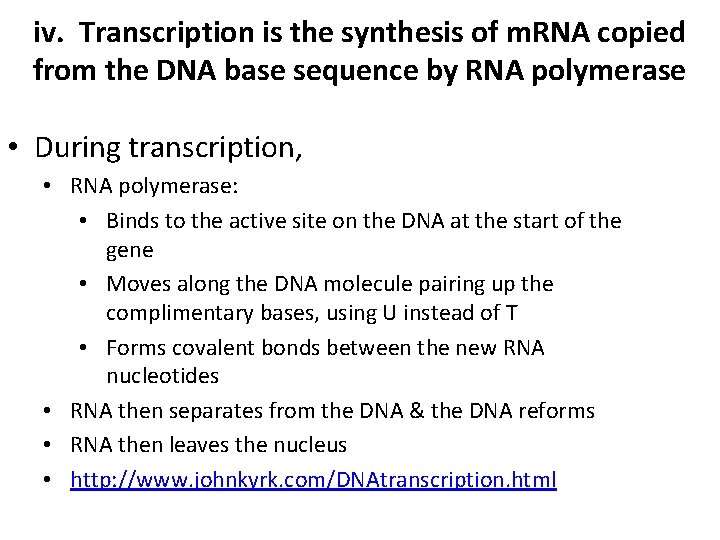
iv. Transcription is the synthesis of m. RNA copied from the DNA base sequence by RNA polymerase • During transcription, • RNA polymerase: • Binds to the active site on the DNA at the start of the gene • Moves along the DNA molecule pairing up the complimentary bases, using U instead of T • Forms covalent bonds between the new RNA nucleotides • RNA then separates from the DNA & the DNA reforms • RNA then leaves the nucleus • http: //www. johnkyrk. com/DNAtranscription. html
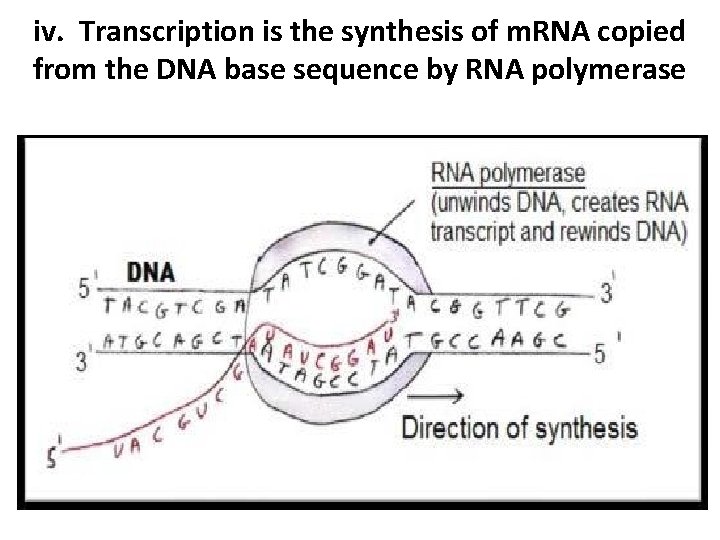
iv. Transcription is the synthesis of m. RNA copied from the DNA base sequence by RNA polymerase
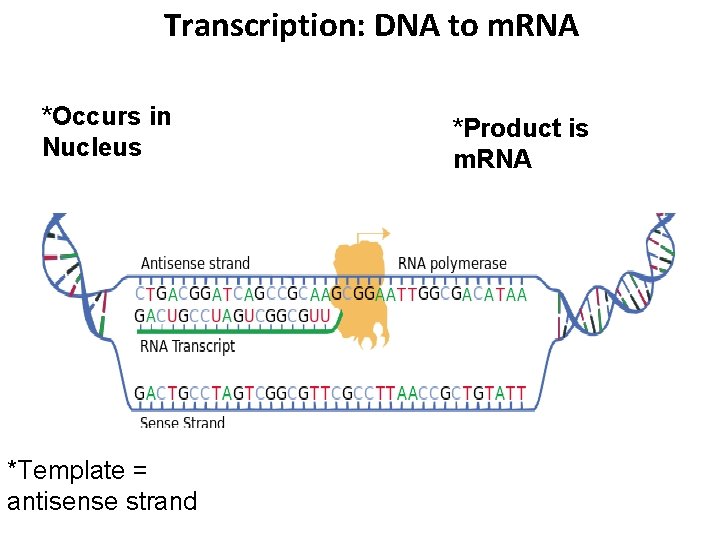
Transcription: DNA to m. RNA *Occurs in Nucleus *Template = antisense strand *Product is m. RNA
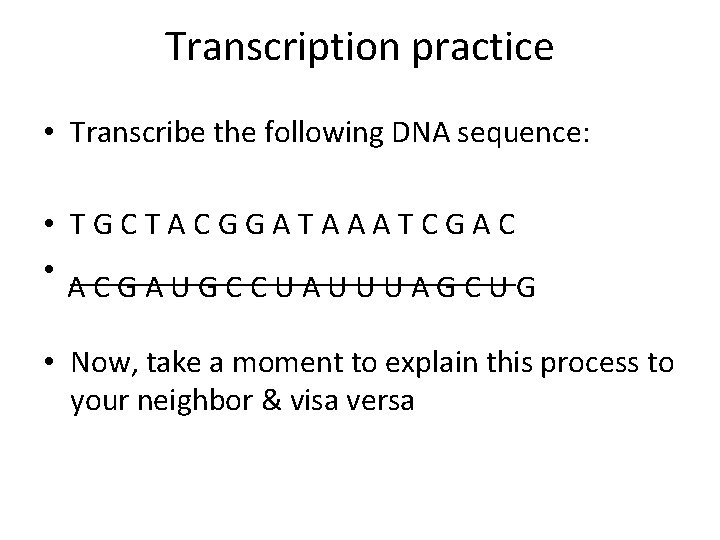
Transcription practice • Transcribe the following DNA sequence: • TGCTACGGATAAATCGAC • ______________ ACGAUGCCUAUUUAGCUG • Now, take a moment to explain this process to your neighbor & visa versa

TRANSLATION: m. RNA to Protein
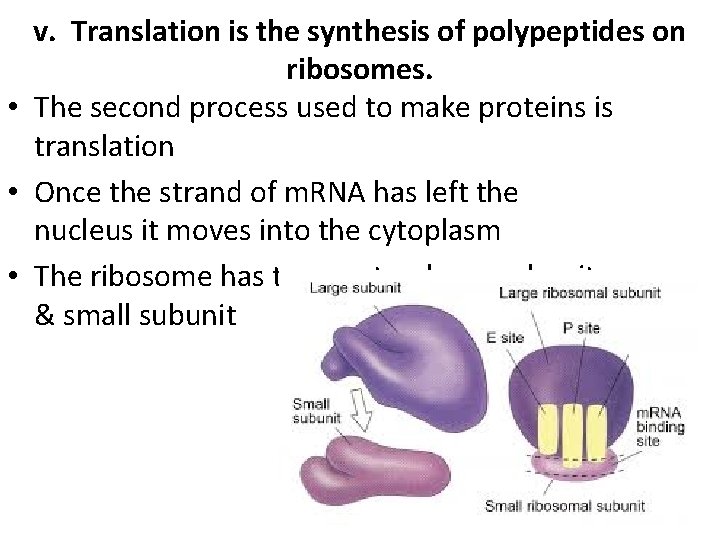
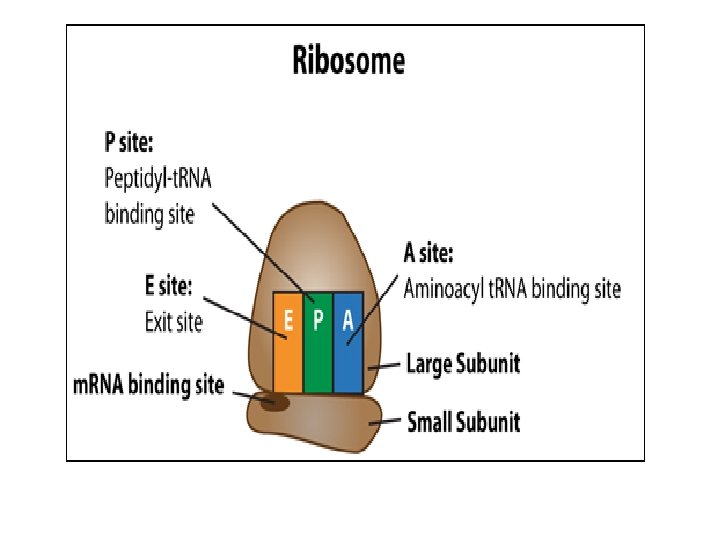
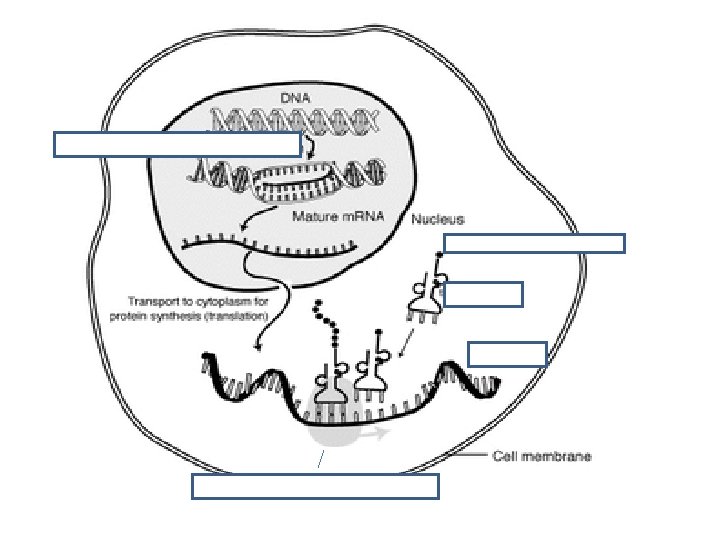
v. Translation is the synthesis of polypeptides on ribosomes. • The second process used to make proteins is translation • Once the strand of m. RNA has left the nucleus it moves into the cytoplasm • The ribosome has two parts – large subunit & small subunit
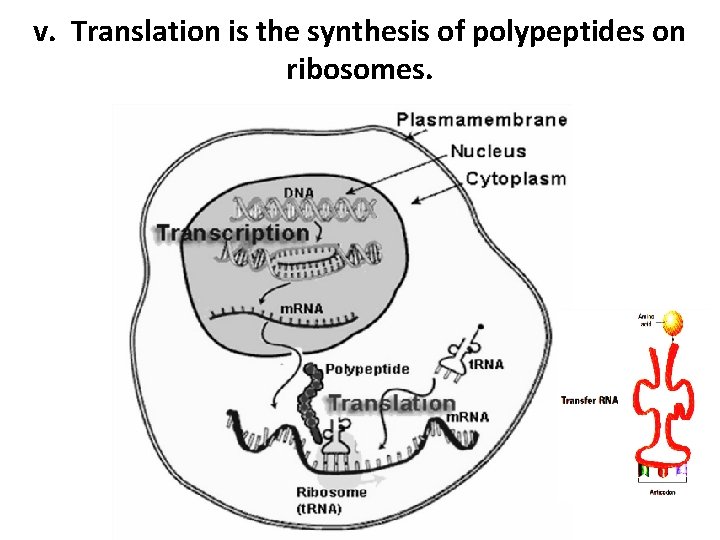
v. Translation is the synthesis of polypeptides on ribosomes.
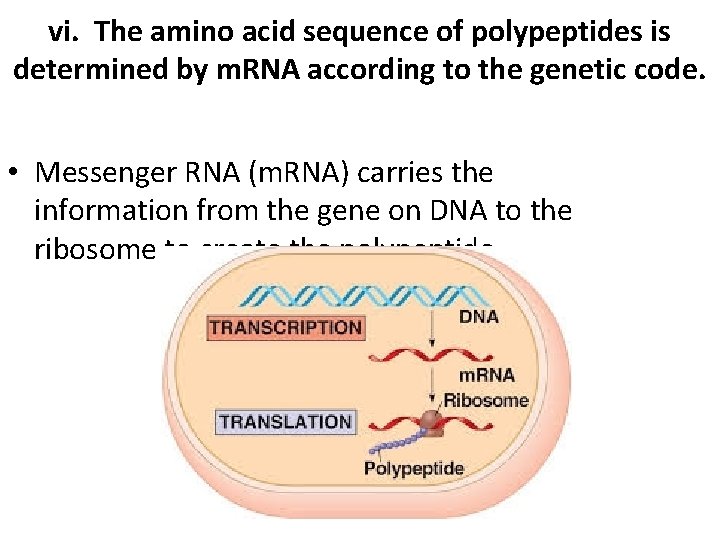
vi. The amino acid sequence of polypeptides is determined by m. RNA according to the genetic code. • Messenger RNA (m. RNA) carries the information from the gene on DNA to the ribosome to create the polypeptide.

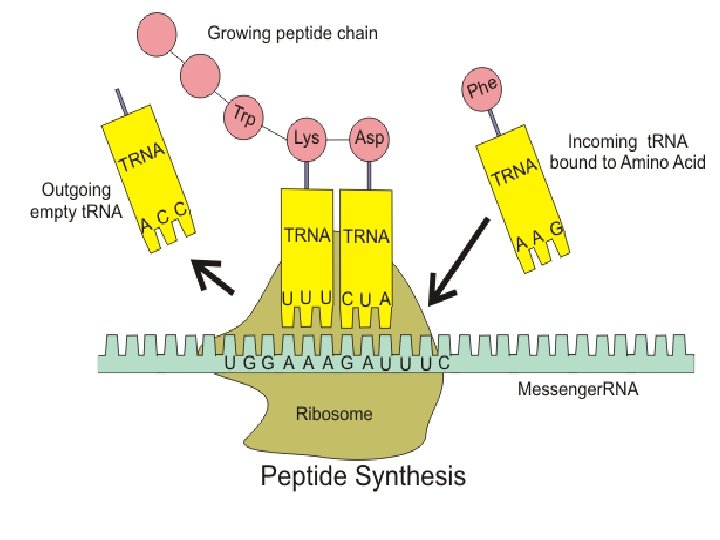
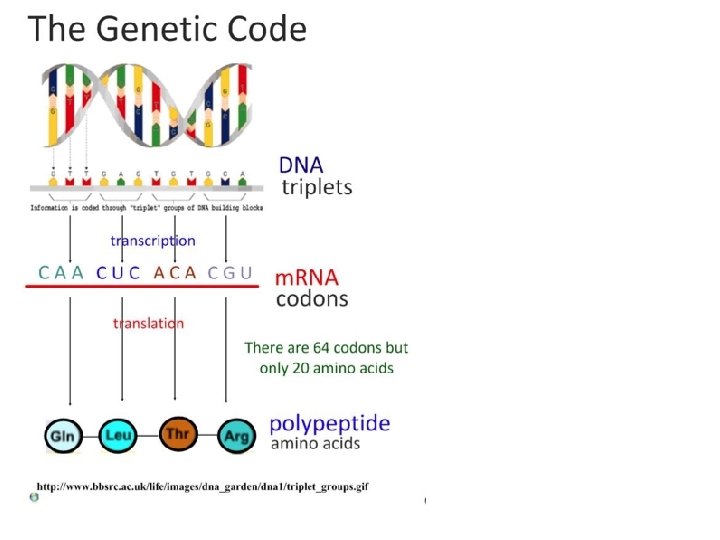
vii. Codons of three bases on m. RNA correspond to one amino acid in a polypeptide • There are 4 different bases & 20 amino acids so a single base cannot code for an amino acid • Every 3 bases therefore code for an amino acid • These groups of 3 bases that are found on the m. RNA is referred to as a codon • Because there are 64 different combinations of codons, several codons code for the same amino acid • When the m. RNA strand pairs up with the t. RNA the 3 bases complimentary to the m. RNA codon is called an anti-codon

vii. Codons of three bases on m. RNA correspond to one amino acid in a polypeptide • 3 components work together to synthesize polypeptides by translation • m. RNA contains codons that code for specific amino acids • t. RNA has a complimentary anticodon on one end that pairs with the codon & an amino acid on the other end • Ribosomes act as a binding site for m. RNA & t. RNA & also catalyze the assembly of the polypeptide

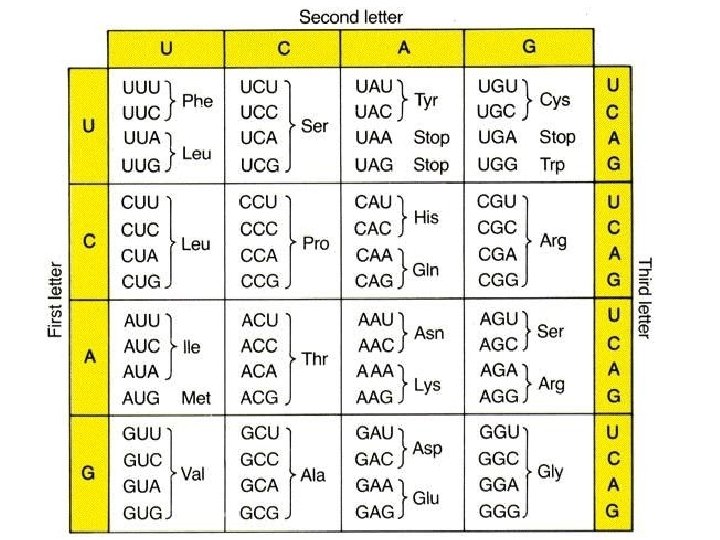
SKILL: Use a table of the genetic code to deduce which codon(s) corresponds to which amino acid.

SKILL: Use a table of m. RNA codons and their corresponding amino acids to deduce the sequence of amino acids coded by a short m. RNA strand of know base sequence

SKILL: Deducing the DNA base sequence for the m. RNA strand
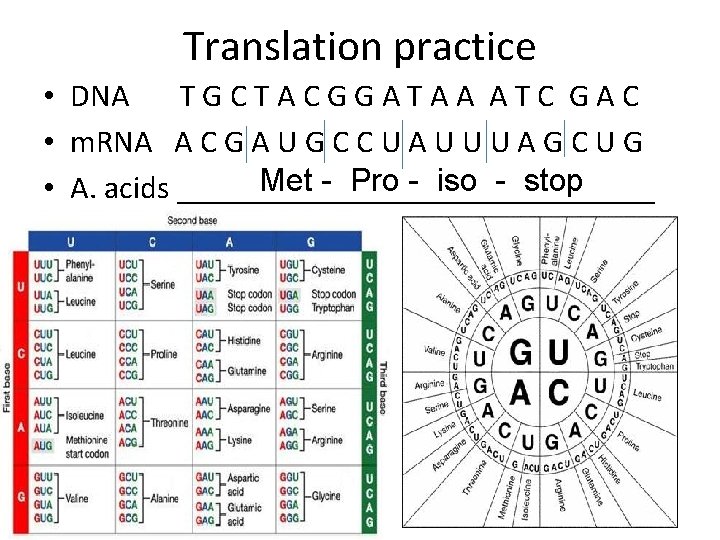
Translation practice • DNA TGCTACGGATAA ATC GAC • m. RNA A C G A U G C C U A U U U A G C U G Met - Pro - iso - stop • A. acids _______________
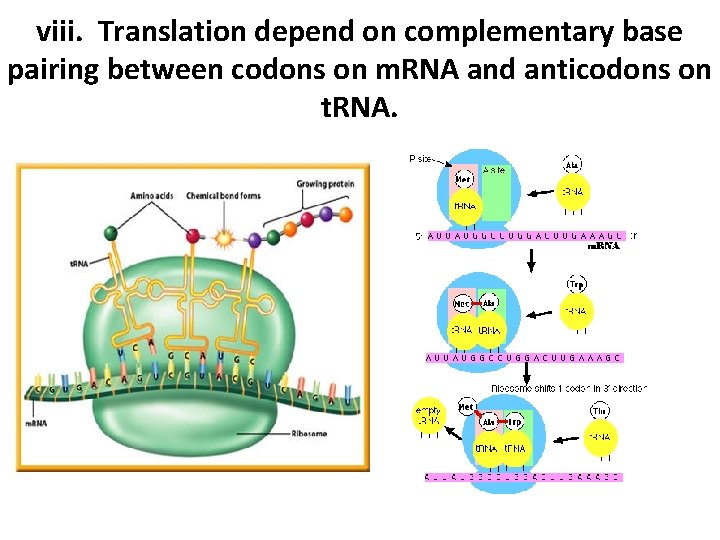
viii. Translation depend on complementary base pairing between codons on m. RNA and anticodons on t. RNA.
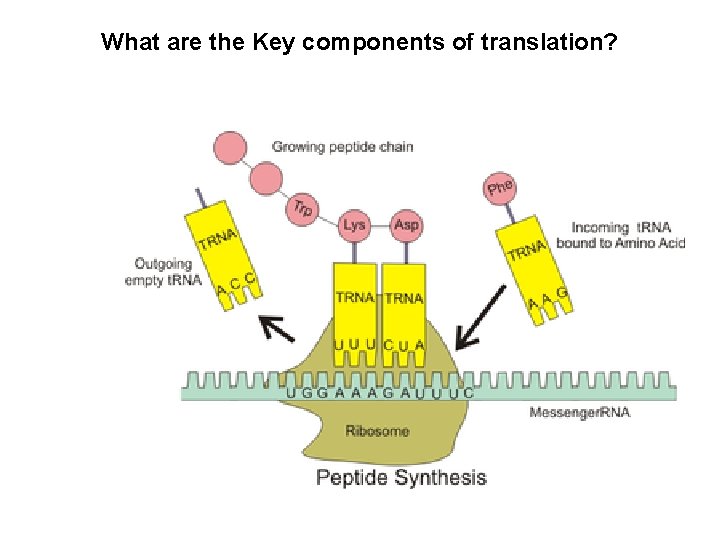
What are the Key components of translation?
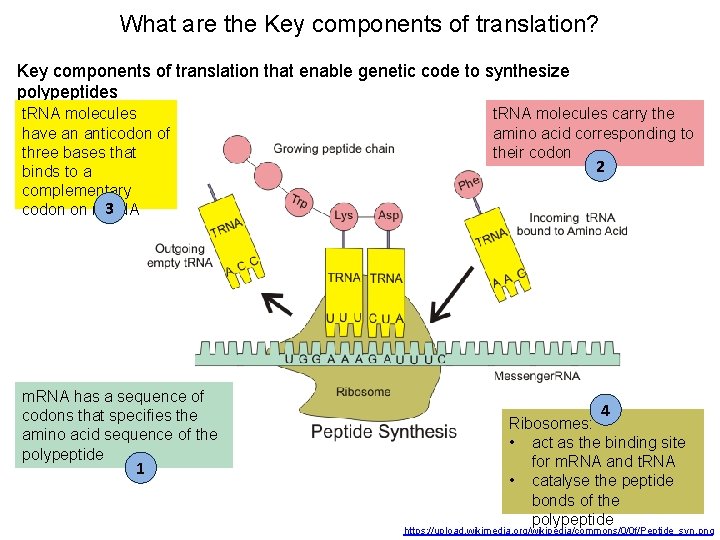
What are the Key components of translation? Key components of translation that enable genetic code to synthesize polypeptides t. RNA molecules have an anticodon of three bases that binds to a complementary 3 codon on m. RNA has a sequence of codons that specifies the amino acid sequence of the polypeptide 1 t. RNA molecules carry the amino acid corresponding to their codon 2 4 Ribosomes: • act as the binding site for m. RNA and t. RNA • catalyse the peptide bonds of the polypeptide https: //upload. wikimedia. org/wikipedia/commons/0/0 f/Peptide_syn. png
viii. Translation depend on complementary base pairing between codons on m. RNA and anticodons on t. RNA. Watch the following animation on translation. http: //highered. mheducation. co m/sites/0072943696/student_view 0/chapte r 3/animation__protein_synthesis__quiz_3_. html
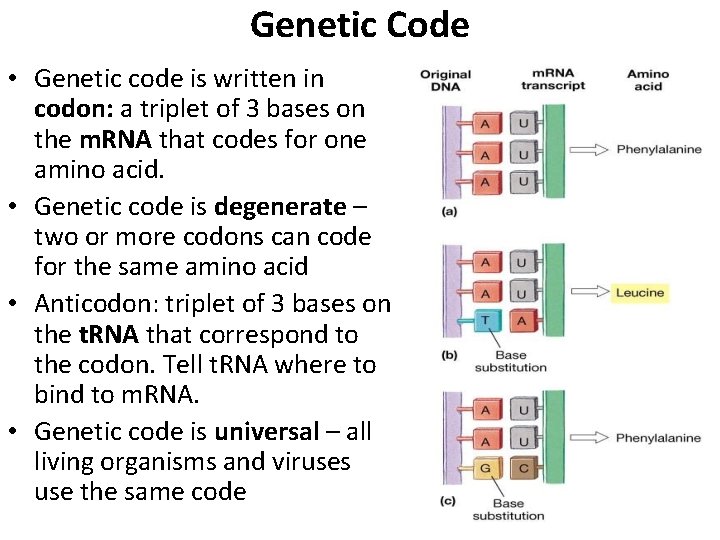
Genetic Code • Genetic code is written in codon: a triplet of 3 bases on the m. RNA that codes for one amino acid. • Genetic code is degenerate – two or more codons can code for the same amino acid • Anticodon: triplet of 3 bases on the t. RNA that correspond to the codon. Tell t. RNA where to bind to m. RNA. • Genetic code is universal – all living organisms and viruses use the same code
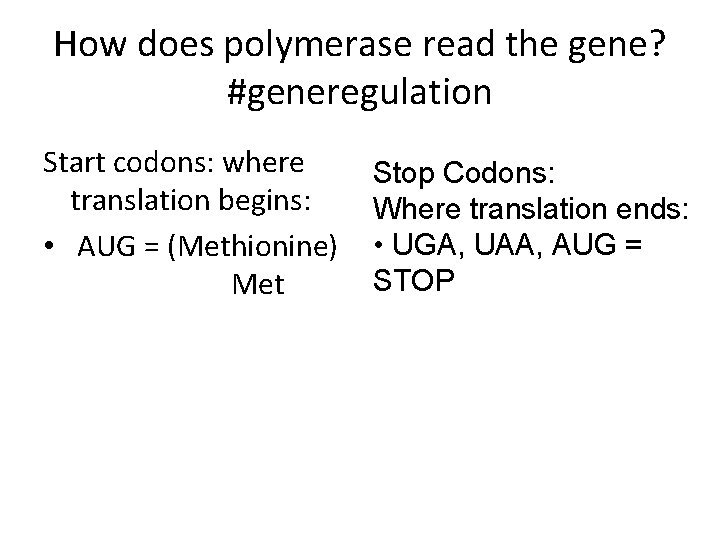
How does polymerase read the gene? #generegulation Start codons: where translation begins: • AUG = (Methionine) Met Stop Codons: Where translation ends: • UGA, UAA, AUG = STOP

Application: Production of human insulin in bacteria as an example of the universality of the genetic code allowing gene transfer between species. Watch the video on the production of insulin and genetic engineering https: //www. youtube. com/watch? v=zlq. D 4 U WCuws 3. 37 min

2. 7. A 2 Production of human insulin in bacteria as an example of the universality of the genetic code allowing gene transfer between species. Diabetes in some individuals is due to destruction of cells in the pancreas that secrete the hormone insulin. It can be treated by injecting insulin into the blood. Porcine and bovine insulin, extracted from the pancreases of pigs and cattle, have both been widely used. Porcine insulin has only one difference in amino acid sequence from human insulin and bovine insulin has three differences. Shark insulin, which has been used for treating diabetics in Japan, has seventeen differences. Despite the differences in the amino acid sequence between animal and human insulin, they all bind to the human insulin receptor and cause lowering of blood glucose concentration. However, some diabetics develop an allergy to animal insulins, so it is preferable to use human insulin. In 1982 human insulin became commercially available for the first time. It was produced using genetically modified E. coli bacteria. Since then methods of production have been developed using yeast cells and more recently safflower plants. https: //en. wikipedia. org/wiki/File: Inzul%C 3%ADn. jpg
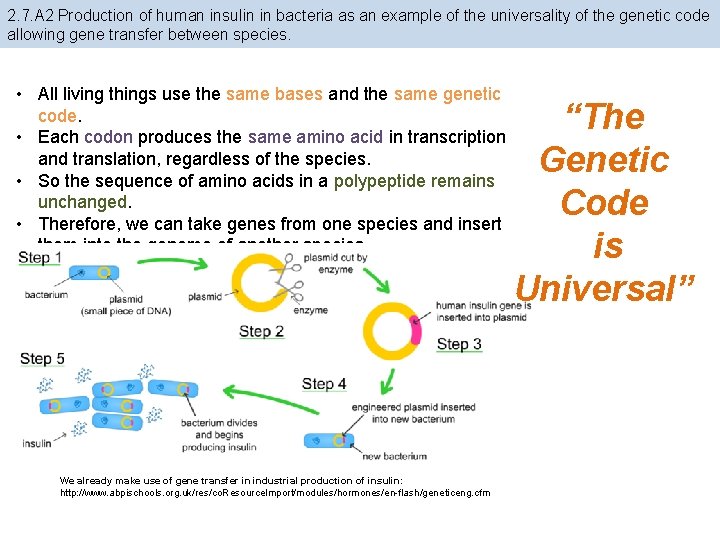
2. 7. A 2 Production of human insulin in bacteria as an example of the universality of the genetic code allowing gene transfer between species. • All living things use the same bases and the same genetic code. • Each codon produces the same amino acid in transcription and translation, regardless of the species. • So the sequence of amino acids in a polypeptide remains unchanged. • Therefore, we can take genes from one species and insert them into the genome of another species. We already make use of gene transfer in industrial production of insulin: http: //www. abpischools. org. uk/res/co. Resource. Import/modules/hormones/en-flash/geneticeng. cfm “The Genetic Code is Universal”
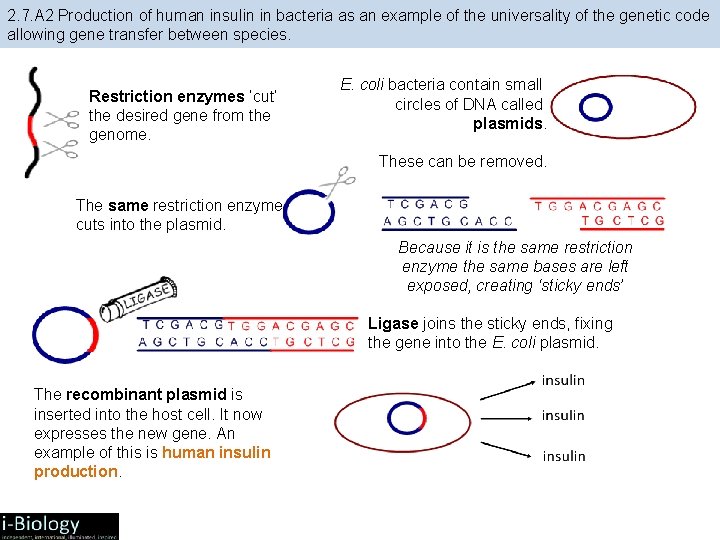
2. 7. A 2 Production of human insulin in bacteria as an example of the universality of the genetic code allowing gene transfer between species. Restriction enzymes ‘cut’ the desired gene from the genome. E. coli bacteria contain small circles of DNA called plasmids. These can be removed. The same restriction enzyme cuts into the plasmid. Because it is the same restriction enzyme the same bases are left exposed, creating ‘sticky ends’ Ligase joins the sticky ends, fixing the gene into the E. coli plasmid. The recombinant plasmid is inserted into the host cell. It now expresses the new gene. An example of this is human insulin production.
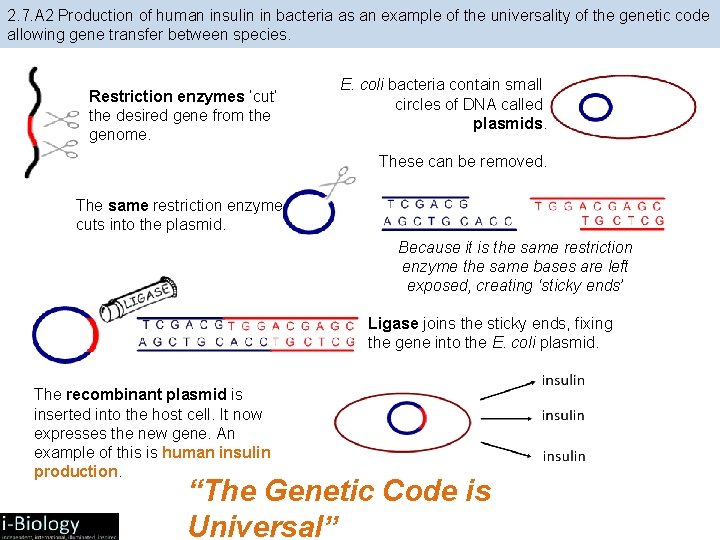
2. 7. A 2 Production of human insulin in bacteria as an example of the universality of the genetic code allowing gene transfer between species. Restriction enzymes ‘cut’ the desired gene from the genome. E. coli bacteria contain small circles of DNA called plasmids. These can be removed. The same restriction enzyme cuts into the plasmid. Because it is the same restriction enzyme the same bases are left exposed, creating ‘sticky ends’ Ligase joins the sticky ends, fixing the gene into the E. coli plasmid. The recombinant plasmid is inserted into the host cell. It now expresses the new gene. An example of this is human insulin production. “The Genetic Code is Universal”
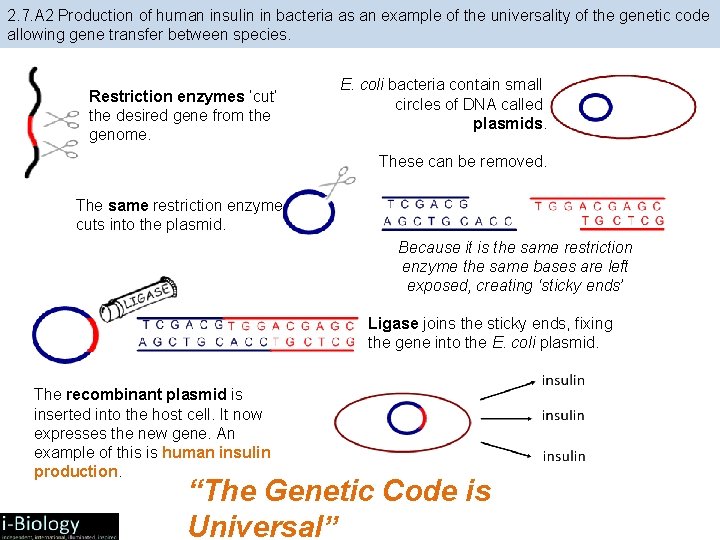
2. 7. A 2 Production of human insulin in bacteria as an example of the universality of the genetic code allowing gene transfer between species. Restriction enzymes ‘cut’ the desired gene from the genome. E. coli bacteria contain small circles of DNA called plasmids. These can be removed. The same restriction enzyme cuts into the plasmid. Because it is the same restriction enzyme the same bases are left exposed, creating ‘sticky ends’ Ligase joins the sticky ends, fixing the gene into the E. coli plasmid. The recombinant plasmid is inserted into the host cell. It now expresses the new gene. An example of this is human insulin production. “The Genetic Code is Universal”
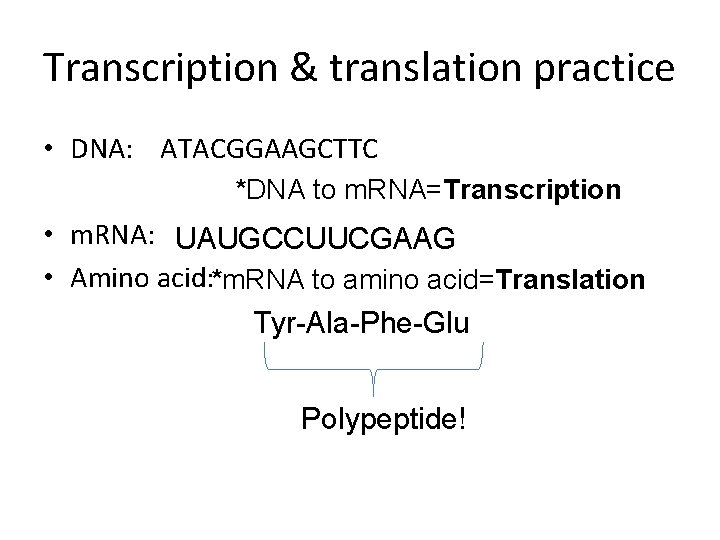
Transcription & translation practice • DNA: ATACGGAAGCTTC *DNA to m. RNA=Transcription • m. RNA: UAUGCCUUCGAAG • Amino acid: *m. RNA to amino acid=Translation Tyr-Ala-Phe-Glu Polypeptide!

Best animation: https: //www. youtube. com/watch? v=D 3 f. OXt 4 Mr. OM Another one: http: //www. youtube. com/watch? v=er. OP 76_q. L WA

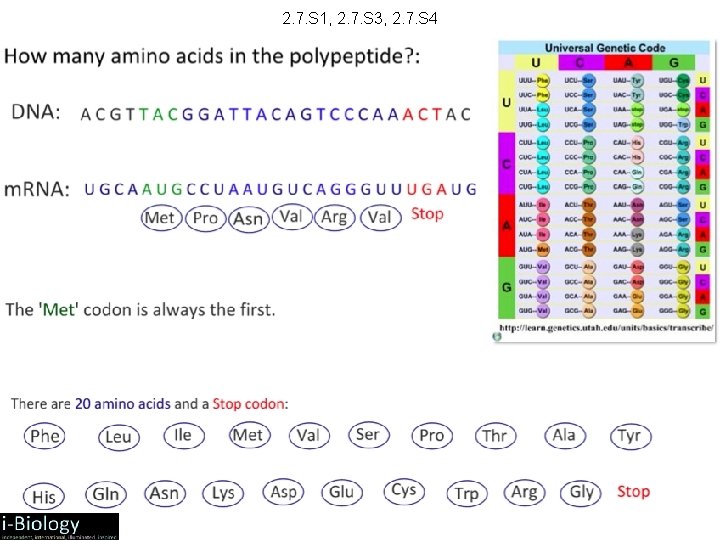
2. 7. S 1, 2. 7. S 3, 2. 7. S 4 1. Deduce the codon(s) that translate for Aspartate. 2. If m. RNA contains the base sequence CUGACUAGGUCCGGA a. deduce the amino acid sequence of the polypeptide translated. b. deduce the base sequence of the DNA antisense strand from which the m. RNA was transcribed. 3. If m. RNA contains the base sequence ACUAAC deduce the base sequence of the DNA sense strand.
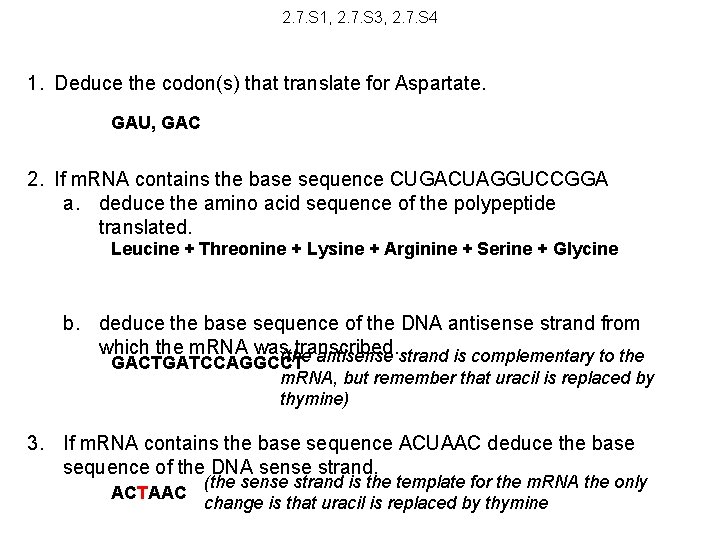
2. 7. S 1, 2. 7. S 3, 2. 7. S 4 1. Deduce the codon(s) that translate for Aspartate. GAU, GAC 2. If m. RNA contains the base sequence CUGACUAGGUCCGGA a. deduce the amino acid sequence of the polypeptide translated. Leucine + Threonine + Lysine + Arginine + Serine + Glycine b. deduce the base sequence of the DNA antisense strand from which the m. RNA was transcribed. (the antisense strand is complementary to the GACTGATCCAGGCCT m. RNA, but remember that uracil is replaced by thymine) 3. If m. RNA contains the base sequence ACUAAC deduce the base sequence of the DNA sense strand. ACTAAC (the sense strand is the template for the m. RNA the only change is that uracil is replaced by thymine

2. 7. S 1, 2. 7. S 3, 2. 7. S 4 1. Deduce the codon(s) that translate for Aspartate. 2. If m. RNA contains the base sequence CUGACUAGGUCCGGA a. deduce the amino acid sequence of the polypeptide translated. b. deduce the base sequence of the DNA antisense strand from which the m. RNA was transcribed. 3. If m. RNA contains the base sequence ACUAAC deduce the base sequence of the DNA sense strand.
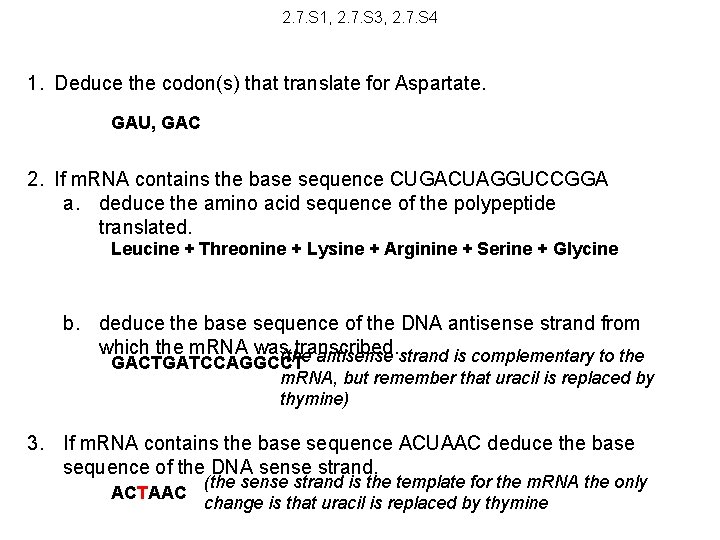
2. 7. S 1, 2. 7. S 3, 2. 7. S 4 1. Deduce the codon(s) that translate for Aspartate. GAU, GAC 2. If m. RNA contains the base sequence CUGACUAGGUCCGGA a. deduce the amino acid sequence of the polypeptide translated. Leucine + Threonine + Lysine + Arginine + Serine + Glycine b. deduce the base sequence of the DNA antisense strand from which the m. RNA was transcribed. (the antisense strand is complementary to the GACTGATCCAGGCCT m. RNA, but remember that uracil is replaced by thymine) 3. If m. RNA contains the base sequence ACUAAC deduce the base sequence of the DNA sense strand. ACTAAC (the sense strand is the template for the m. RNA the only change is that uracil is replaced by thymine

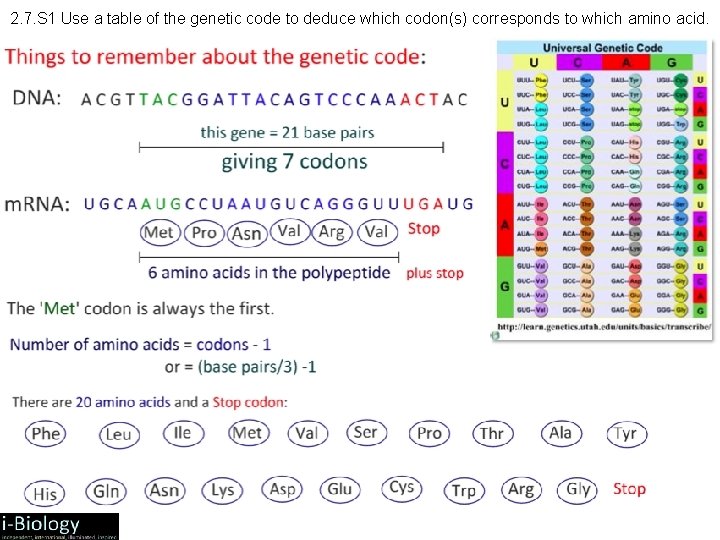
2. 7. S 1 Use a table of the genetic code to deduce which codon(s) corresponds to which amino acid.

2. 7. S 1, 2. 7. S 3, 2. 7. S 4
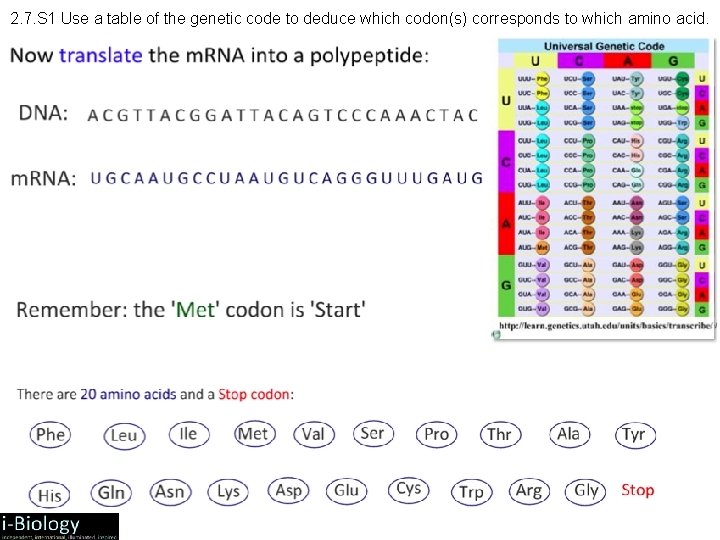
2. 7. S 1 Use a table of the genetic code to deduce which codon(s) corresponds to which amino acid.
2. 7. S 1, 2. 7. S 3, 2. 7. S 4
2. 7. S 1 Use a table of the genetic code to deduce which codon(s) corresponds to which amino acid.
- Slides: 71