Topic 10 2 INHERITANCE Topic Outline Genetic Organisation




- Slides: 20

Topic 10. 2 INHERITANCE

Topic Outline þ Genetic Organisation þ Mendelian Inheritance Patterns þ Monohybrid versus Dihybrid Crosses þ Non-Mendelian Inheritance Patterns þ Gene Linkage þ Recombination þ Polygenic Inheritance Genes may be linked or unlinked and are inherited accordingly AUDIO

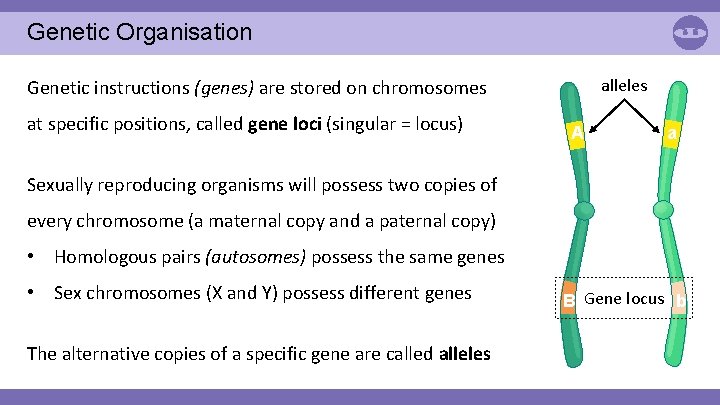
Genetic Organisation alleles Genetic instructions (genes) are stored on chromosomes at specific positions, called gene loci (singular = locus) A a Sexually reproducing organisms will possess two copies of every chromosome (a maternal copy and a paternal copy) • Homologous pairs (autosomes) possess the same genes • Sex chromosomes (X and Y) possess different genes The alternative copies of a specific gene are called alleles B Gene locus b

Part 1: MENDELIAN INHERITANCE

Mendel Gregor Mendel was an Austrian monk who developed the principles of inheritance by experimenting on pea plants • Law of Segregation: All sexually reproducing organisms inherit one copy of a gene (i. e. allele) from each parent • Law of Independent Assortment: Inheritance of alleles for one gene occurs independently of all other genes* * Not true for genes located on the same chromosome (linked) Gregor Mendel (1822 – 1884)

Mendelian Inheritance Summary: • Gregor Mendel • Pea experiments • Genotypes & alleles • Dominance hierarchy • Punnett squares • Monohybrid crosses • Dihybrid crosses Video outlining principles of Mendelian inheritance

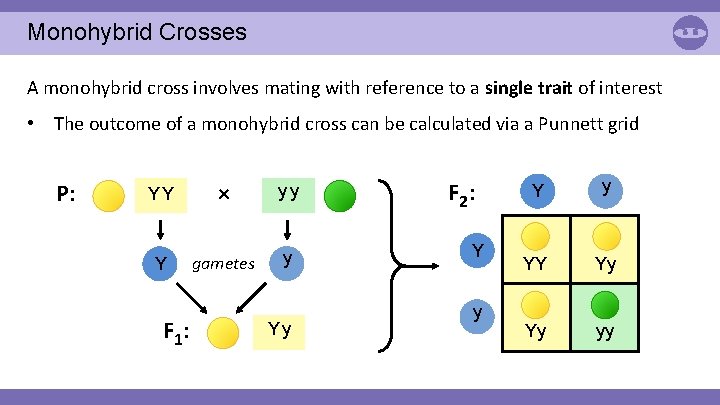
Monohybrid Crosses A monohybrid cross involves mating with reference to a single trait of interest • The outcome of a monohybrid cross can be calculated via a Punnett grid P: YY × yy Y gametes y F 1: Yy F 2: Y y YY Yy Yy yy

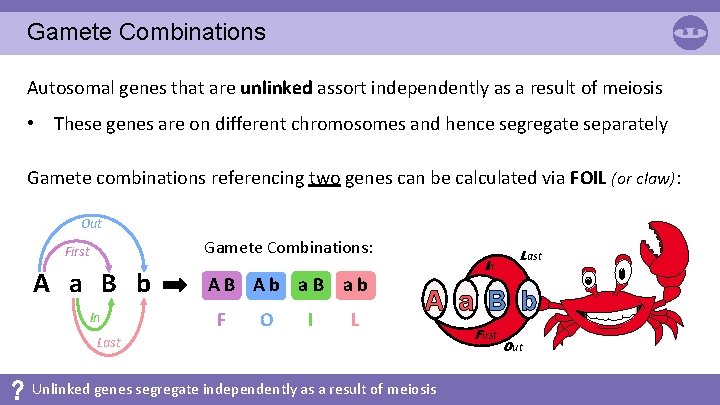
Gamete Combinations Autosomal genes that are unlinked assort independently as a result of meiosis • These genes are on different chromosomes and hence segregate separately Gamete combinations referencing two genes can be calculated via FOIL (or claw): Out Gamete Combinations: First A a B b In AB Ab a. B ab F O I L In Last A a B b Last Unlinked genes segregate independently as a result of meiosis First Out

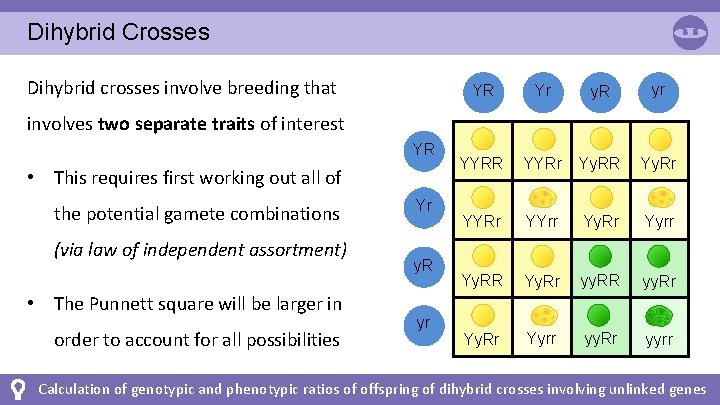
Dihybrid Crosses Dihybrid crosses involve breeding that YR Yr y. R yr YYRR YYRr Yy. RR Yy. Rr YYrr Yy. Rr Yyrr Yy. RR Yy. Rr yy. RR yy. Rr Yyrr yy. Rr yyrr involves two separate traits of interest YR • This requires first working out all of the potential gamete combinations (via law of independent assortment) • The Punnett square will be larger in order to account for all possibilities Yr y. R yr Calculation of genotypic and phenotypic ratios of offspring of dihybrid crosses involving unlinked genes

Part 2: NON-MENDELIAN INHERITANCE

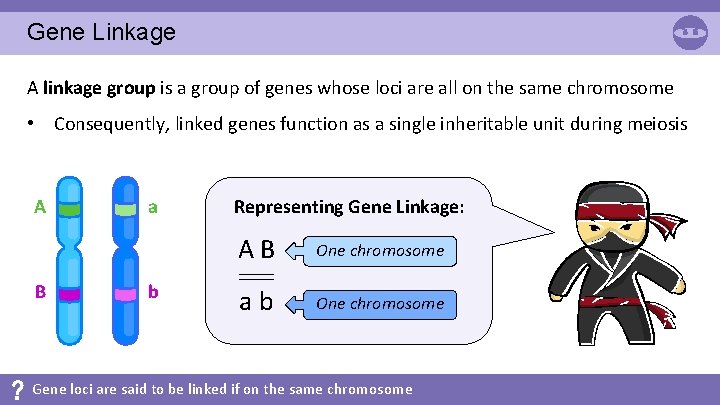
Gene Linkage A linkage group is a group of genes whose loci are all on the same chromosome • Consequently, linked genes function as a single inheritable unit during meiosis A B a b Representing Gene Linkage: AB One chromosome ab One chromosome Gene loci are said to be linked if on the same chromosome

Thomas Morgan developed the concept of gene linkage via breeding experiments with fruit flies (Drosophila) He found a sex bias in the inheritance of certain key traits • Eye color (red vs white), wing length (normal vs vestigial) He deduced that genes located on a shared chromosome (e. g. chromosome 2) would not assort independently • These genes show non-Mendelian inheritance ratios Morgan’s discovery of non-Mendelian ratios in Drosophila Thomas Morgan

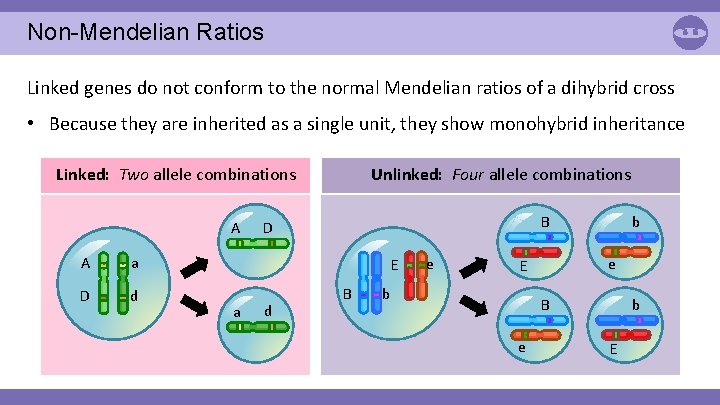
Non-Mendelian Ratios Linked genes do not conform to the normal Mendelian ratios of a dihybrid cross • Because they are inherited as a single unit, they show monohybrid inheritance Linked: Two allele combinations A A a D d Unlinked: Four allele combinations E a d b B D B e e E b b B e E

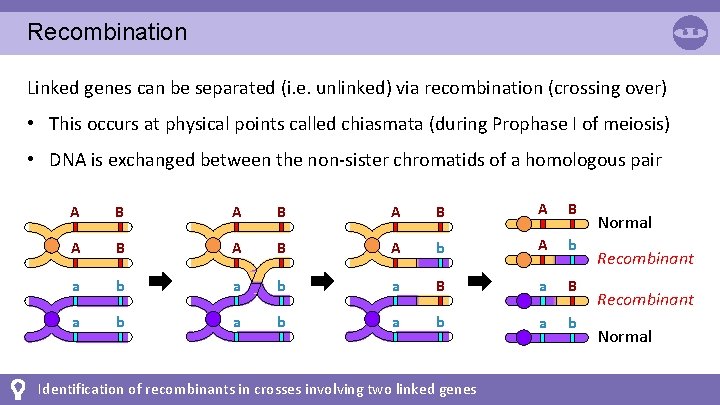
Recombination Linked genes can be separated (i. e. unlinked) via recombination (crossing over) • This occurs at physical points called chiasmata (during Prophase I of meiosis) • DNA is exchanged between the non-sister chromatids of a homologous pair A B A B A B A b a b a B a b a b Identification of recombinants in crosses involving two linked genes Normal Recombinant Normal

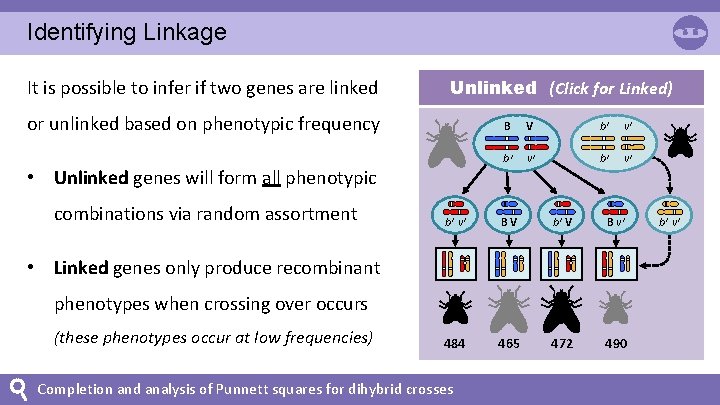
Identifying Linkage It is possible to infer if two genes are linked Unlinked (Click for Linked) or unlinked based on phenotypic frequency • Unlinked genes will form all phenotypic combinations via random assortment B V b+ v+ BV b+ V B v+ 484 465 472 490 • Linked genes only produce recombinant phenotypes when crossing over occurs (these phenotypes occur at low frequencies) Completion and analysis of Punnett squares for dihybrid crosses b+ v+

Part 3: POLYGENIC INHERITANCE

Phenotypic Variation Different allele combinations result in divergent phenotypic characteristics • As more genes contribute to the expression of a trait, the variation increases • Variation can be discrete (specific phenotypes) or continuous (along a spectrum) Monogenic traits are controlled by a single gene and show discrete variation • Examples of monogenic traits include presence of attached earlobes / freckles Polygenic traits are controlled by multiple genes and show continuous variation • Examples of polygenic traits include human height, weight and skin colour Variation can be continuous or discrete

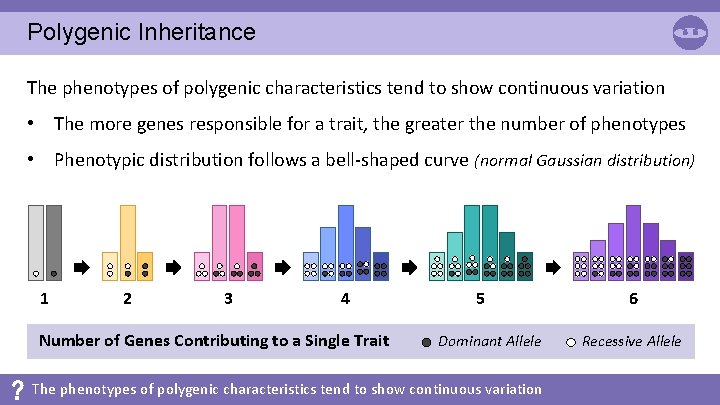
Polygenic Inheritance The phenotypes of polygenic characteristics tend to show continuous variation • The more genes responsible for a trait, the greater the number of phenotypes • Phenotypic distribution follows a bell-shaped curve (normal Gaussian distribution) 1 2 3 4 Number of Genes Contributing to a Single Trait 5 Dominant Allele The phenotypes of polygenic characteristics tend to show continuous variation 6 Recessive Allele

Polygenic Examples Polygenic traits are determined by genotype and also by environmental factors Human Height • Controlled by multiple genes, resulting in a bell-shaped phenotype spectrum • Environmental factors such as diet and health (i. e. disease) can influence height Human Skin Colour • Overall skin pigmentation is controlled by multiple melanin producing genes • It is also impacted by external factors such as sun exposure or vitamin D levels Polygenic traits such as human height may also be influenced by environmental factors

Topic Review Can you do the following? • Outline how genetic information is organised • Calculate dihybrid ratios from provided data • Describe gene linkage • Identify recombinant phenotypes • Compare linked and unlinked inheritance ratios • Distinguish between types of variation • Identify examples of polygenic inheritance