Time to CARE A collaborative engine for practical

Time to CARE: A collaborative engine for practical disease prediction D. Davis et al. (2009) in Data mining and knowledge discovery Speaker: Sang Ho Oh Feb. 20 th on 2018

Introduction • Annual health care expenditure in the U. S. alone is an overwhelming sum. – Majority of this money is used for disease treatment. • Experts expect the burden on the medical system to continually increase in coming years. – In 2001, 3. 1 visits/patient were made to physician. • In history, researchers shown many conditions to have recognizable indicators before onset/preventable risk factors. – The prospective medicine and aim at minimizing the risk can be done. Current situation: • Physicians can use family and health history and physical examination to approximate the risk of patient. • Medical care is reactive, stepping in once the symptoms have emerged. How to prevent? • Prevailing model of prospective health care -> Genome revolution. – Not yet matured. Then what is the option? • Phenotype and disease history based approaches offer the promise of advances towards disease prediction. 2

Purpose of the study Aim of the study: • Development of a predictive system (called CARE: Collaborative Assessment and Recommendation Engine). – How? Examining the use of medical history – For? To examine information about disease correlations and inexpensively assess risk. • How to predict about the future diseases a patient may develop? – Generate a patient’s prognosis based on the experiences of other similar patients. Method used in the study: • Collaborative filtering (will be explained in next page). Contributions of the study: • A novel application of collaborative filtering in the medical domain for advancing the field of prospective medicine. • Present a general system which makes predictions on all types of diseases and medical conditions (using ICD-9 -CM). *ICD-9 -CM: International classification of diseases codes. 3

Collaborative filtering • It is designed to predict the preferences of one person(active user) based on the preferences of other similar persons(users). – Assumption: people will enjoy the same items as their similar peers. • Having some common preferences is a strong predictor of additional common preferences. • • Predictions are based on datasets consisting of many user profiles Accomplished by calculating a weight of similarity between active user and all others. – Active user’s opinion is determined by the weighted average of the others’ opinion. How is it applied in medical area? • Each user is a patients whose profile is a diagnosed disease. • Using collaborative filtering, they generated predictions on other diseases based on a set of other similar patients. Difference between original and modified version of collaborative filtering • The rating is binary: either patient has a disease (1) or not (0). 4
Data used • • • The database comprises the Medicare records of 13, 039, 018 elderly patients in U. S. with total of 32, 341, 348 visits. The input for the methods consists of each patient’s diagnosis history and provided per inpatient visit. Each data record consists of hospital visit, patient ID, and list of up to 10 diagnosis codes per visit. – The diagnosis code – International Classification of Diseases, 9 th revision, Clinical Modification (ICD-9 -CM). – Each disease is given a unique code that can be up to 5 character long. – ICD-9 codes are hierarchical in nature so it can be collapsed to fewer characters which identifies a small family of related medical conditions. – There are total of 18, 207 unique disease codes expressed. *Example of collapsing code 40201 - malignant hypertensive heart disease with heart failure. 4020 – non-specific malignant hypertensive heart disease. 402 - family of all hypertensive heart disease. 5
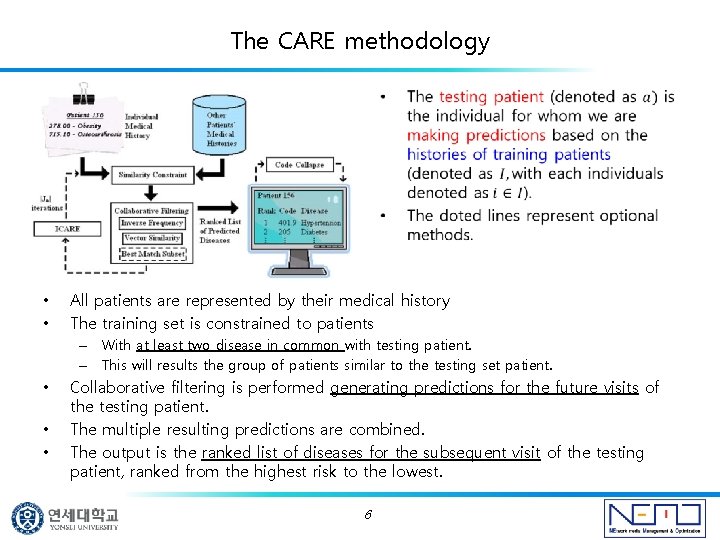
The CARE methodology • • All patients are represented by their medical history The training set is constrained to patients – With at least two disease in common with testing patient. – This will results the group of patients similar to the testing set patient. • • • Collaborative filtering is performed generating predictions for the future visits of the testing patient. The multiple resulting predictions are combined. The output is the ranked list of diseases for the subsequent visit of the testing patient, ranked from the highest risk to the lowest. 6
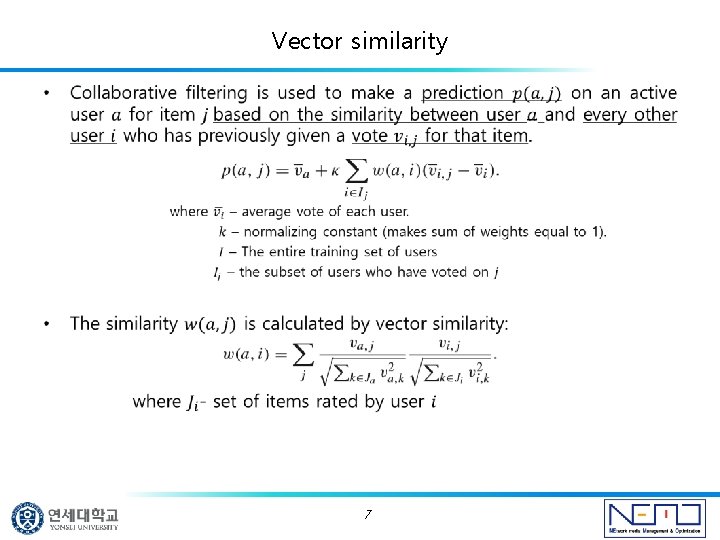
Vector similarity • 7
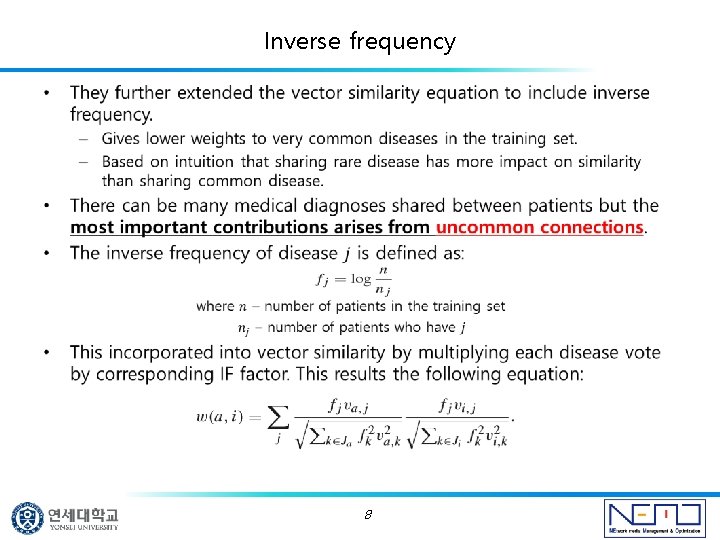
Inverse frequency • 8

Grouping of training patients • Before application of collaborative filtering, a group of relevant training patients is determined. – Based on the number of diagnoses in common with the testing patient. Why? • To remove the influence of patients who have little or no similarity. • Training patients with no disease in common with the active patient do not contribute to the prediction score. • Removing those does not result in loss of information but effectively reduces the runtime of the algorithm. How it works in CARE? • In CARE, they include all patients with 2 or more diseases in common. • This constraint enforces stronger similarities for all patients influencing the predictions. • Helps to avoid the noise. 9
Optional methods • ICARE – This means the “Iterative CARE” – This method developed to capture the effect of each individual disease with minimal noise from other diseases but without loss of information due to removing them. • ICD-9 -CM code collapse – In some cases, it is desirable for 4/5 digit ICD-9 -CM codes to be collapsed in to more general 3 digit code which represent small groups of related/similar disease. – There are two method: truncated to 3 -digits before (pre-collapse) or after (post-collapse) applying collaborative filtering. • Pre-collapse: significantly reduces the runtime of algorithm. • Post-collapse: makes the result simpler to evaluate and interpret. • Time-sensitive CARE – CARE & ICARE do not take the order of or length between disease diagnoses when generating vector similarity. – But matching with two diseases which occurred many years apart may not be relevant. – For that reason, they modified the method to incorporate the length of time between medical event. 10
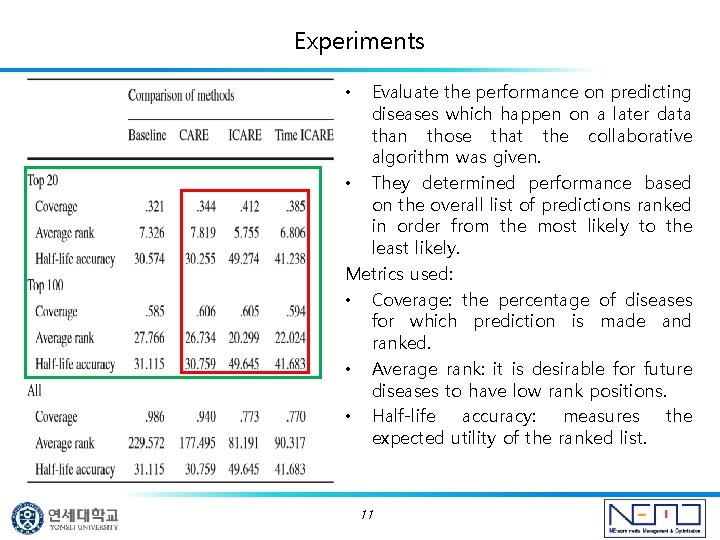
Experiments Evaluate the performance on predicting diseases which happen on a later data than those that the collaborative algorithm was given. • They determined performance based on the overall list of predictions ranked in order from the most likely to the least likely. Metrics used: • Coverage: the percentage of diseases for which prediction is made and ranked. • Average rank: it is desirable for future diseases to have low rank positions. • Half-life accuracy: measures the expected utility of the ranked list. • 11
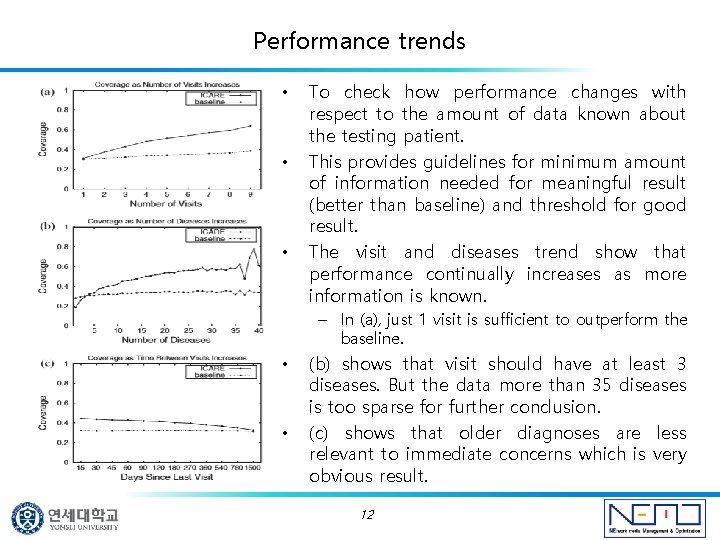
Performance trends • • • To check how performance changes with respect to the amount of data known about the testing patient. This provides guidelines for minimum amount of information needed for meaningful result (better than baseline) and threshold for good result. The visit and diseases trend show that performance continually increases as more information is known. – In (a), just 1 visit is sufficient to outperform the baseline. • • (b) shows that visit should have at least 3 diseases. But the data more than 35 diseases is too sparse for further conclusion. (c) shows that older diagnoses are less relevant to immediate concerns which is very obvious result. 12

Conclusions • • • The goal of the paper is to come up with a system that can assist a medical practitioner in decision making. The authors proposed CARE, a collaborative recommendation engine for prospective and proactive healthcare. This CARE, ICARE, and time-sensitive CARE can predict and provide the future diagnoses of the patient to doctor. – then appropriate medical test can be proceeded. – Improves the quality of life for the patient. – Also can reduce the health care costs. 13

Thank you 14
- Slides: 14