Three Point Mapping Experiment Backward Analysis In a
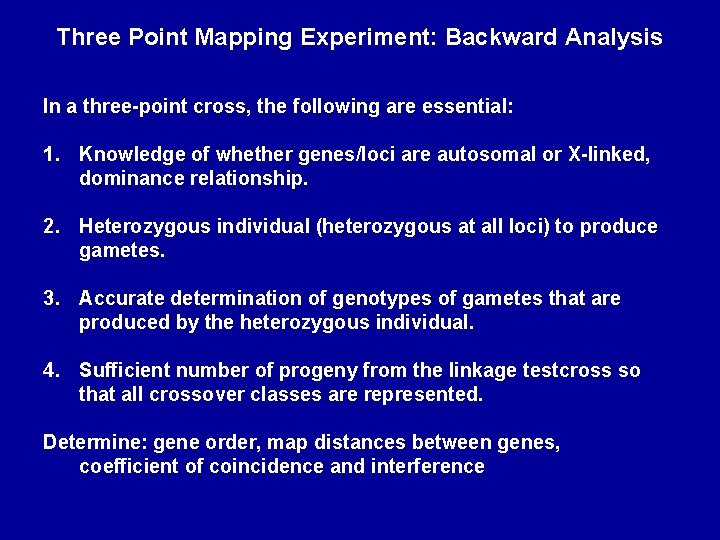
Three Point Mapping Experiment: Backward Analysis In a three-point cross, the following are essential: 1. Knowledge of whether genes/loci are autosomal or X-linked, dominance relationship. 2. Heterozygous individual (heterozygous at all loci) to produce gametes. 3. Accurate determination of genotypes of gametes that are produced by the heterozygous individual. 4. Sufficient number of progeny from the linkage testcross so that all crossover classes are represented. Determine: gene order, map distances between genes, coefficient of coincidence and interference
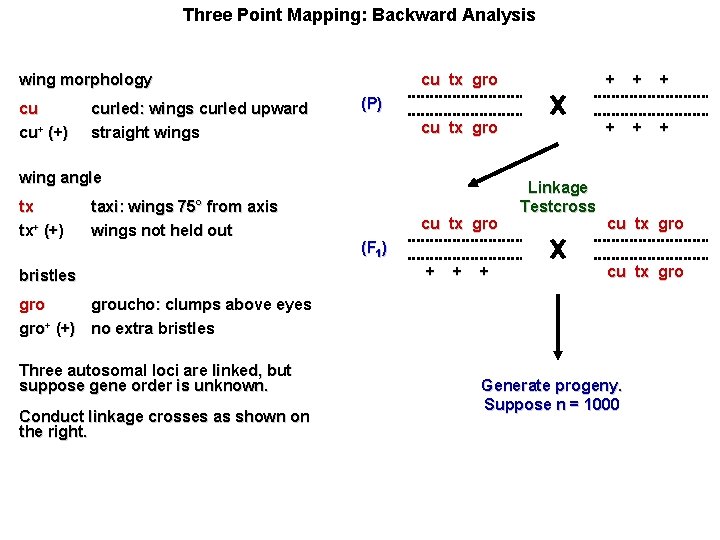
Three Point Mapping: Backward Analysis wing morphology cu cu+ (+) curled: wings curled upward straight wings cu tx gro + + + (P) wing angle tx tx+ (+) taxi: wings 75° from axis wings not held out bristles cu tx gro Linkage Testcross cu tx gro (F 1) + + + cu tx gro groucho: clumps above eyes gro+ (+) no extra bristles Three autosomal loci are linked, but suppose gene order is unknown. Conduct linkage crosses as shown on the right. Generate progeny. Suppose n = 1000
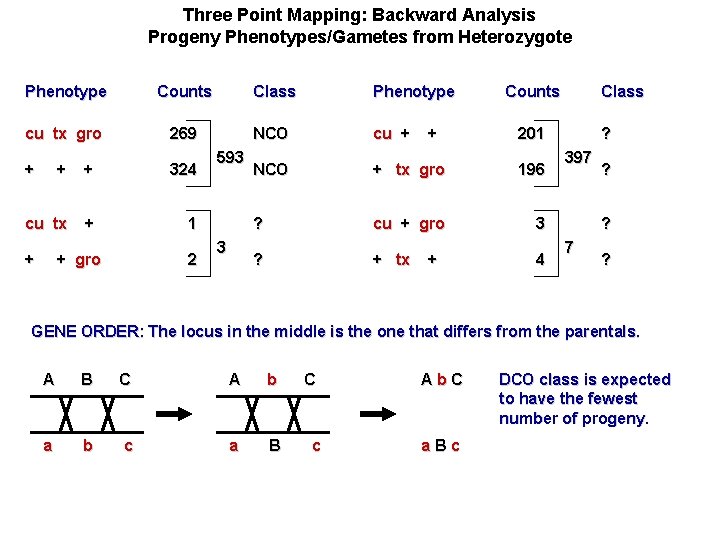
Three Point Mapping: Backward Analysis Progeny Phenotypes/Gametes from Heterozygote Phenotype Counts Class Phenotype cu tx gro 269 NCO cu + + NCO + tx gro 196 ? cu + gro 3 ? + tx 4 + + 324 cu tx + 1 + + gro 2 593 3 + + Counts Class 201 ? 397 ? ? 7 ? GENE ORDER: The locus in the middle is the one that differs from the parentals. A B C A b a b c a B C c Ab. C a. Bc DCO class is expected to have the fewest number of progeny.
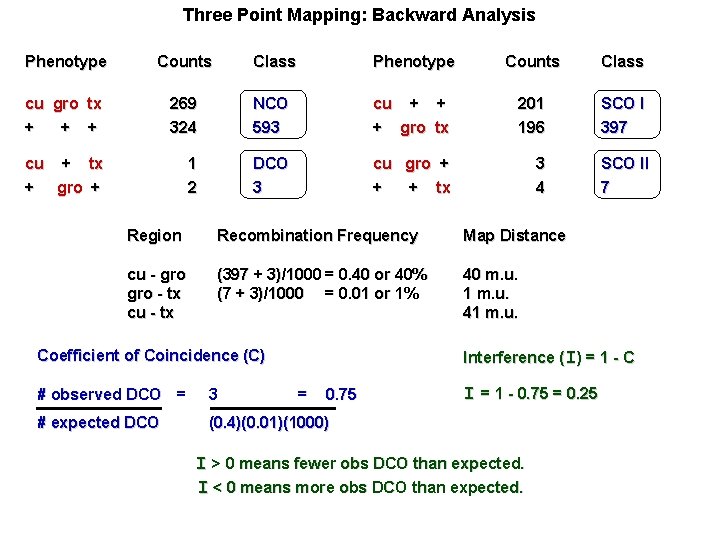
Three Point Mapping: Backward Analysis Phenotype Counts Class cu gro tx + + + 269 324 NCO 593 cu + + + gro tx 201 196 SCO I 397 cu + tx + gro + 1 2 DCO 3 cu gro + + + tx 3 4 SCO II 7 Region Recombination Frequency Map Distance cu - gro - tx cu - tx (397 + 3)/1000 = 0. 40 or 40% (7 + 3)/1000 = 0. 01 or 1% 40 m. u. 1 m. u. 41 m. u. Interference (I) = 1 - C Coefficient of Coincidence (C) # observed DCO = 3 = 0. 75 # expected DCO (0. 4)(0. 01)(1000) I = 1 - 0. 75 = 0. 25 I > 0 means fewer obs DCO than expected. I < 0 means more obs DCO than expected.
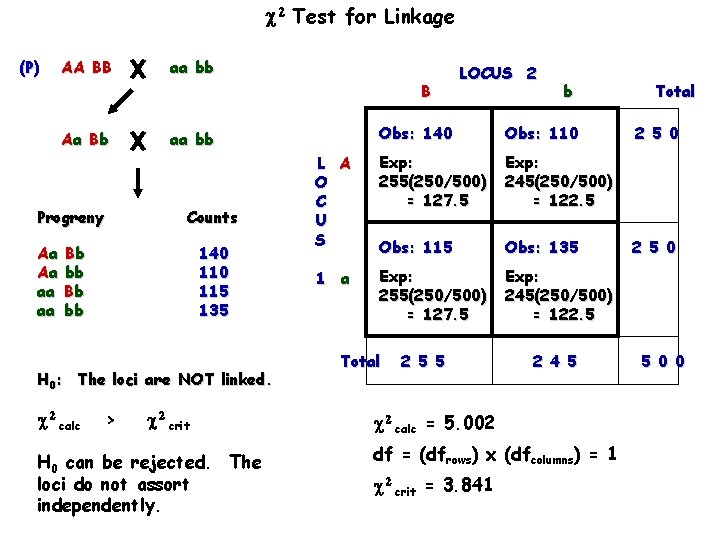
Test for Linkage (P) AA BB aa bb B Aa B b Progreny Aa Aa aa aa Bb bb 140 115 135 H 0: The loci are NOT linked. calc > crit H 0 can be rejected. The loci do not assort independently. b Obs: 140 Obs: 110 L A O C U S Exp: 255(250/500) = 127. 5 Exp: 245(250/500) = 122. 5 Obs: 115 Obs: 135 1 a Exp: 255(250/500) = 127. 5 Exp: 245(250/500) = 122. 5 aa bb Counts LOCUS 2 Total 2 5 5 2 4 5 calc = 5. 002 df = (dfrows) x (dfcolumns) = 1 crit = 3. 841 Total 2 5 0 5 0 0
- Slides: 5