Theoretical and Experimental Experimental and Theoretical description of









![How to determine peptide affinity Law of mass action koff [R] + [L] kon How to determine peptide affinity Law of mass action koff [R] + [L] kon](https://slidetodoc.com/presentation_image/109932c5e1f17507c80f339796b20e83/image-25.jpg)




- Slides: 36

“Theoretical and Experimental “Experimental and Theoretical description of Peptide-MHC binding” Christina Sylvester-Hvid, IMMI, Panum

Generation of Recombinant MHC Class I and Characterization of the People Interaction with Peptide - Contributions to the Human MHC project Christina Sylvester-Hvid, IMMI, Panum

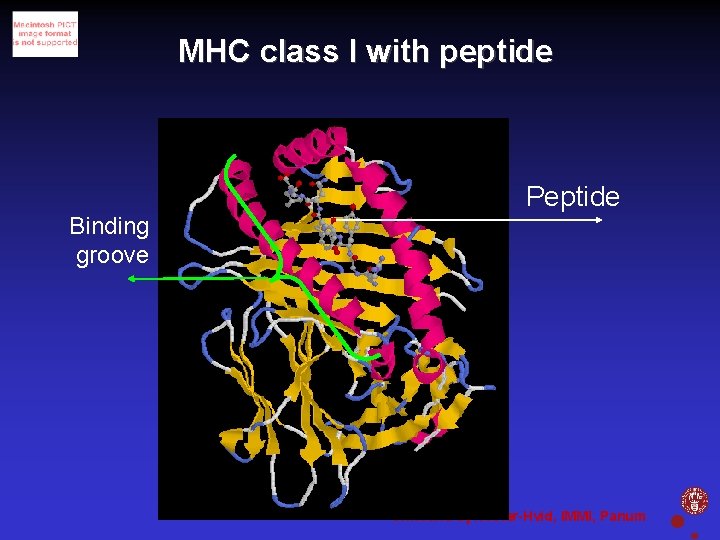
MHC class I with peptide Peptide Binding groove Christina Sylvester-Hvid, IMMI, Panum

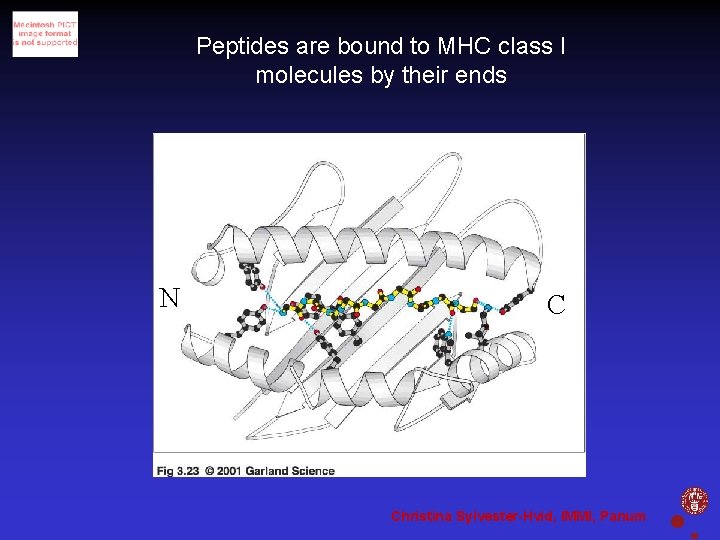
Peptides are bound to MHC class I molecules by their ends N C Christina Sylvester-Hvid, IMMI, Panum

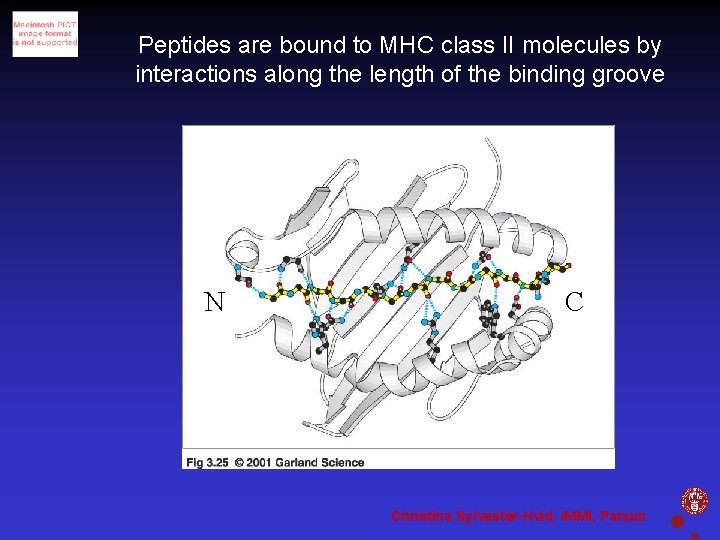
Peptides are bound to MHC class II molecules by interactions along the length of the binding groove N C Christina Sylvester-Hvid, IMMI, Panum

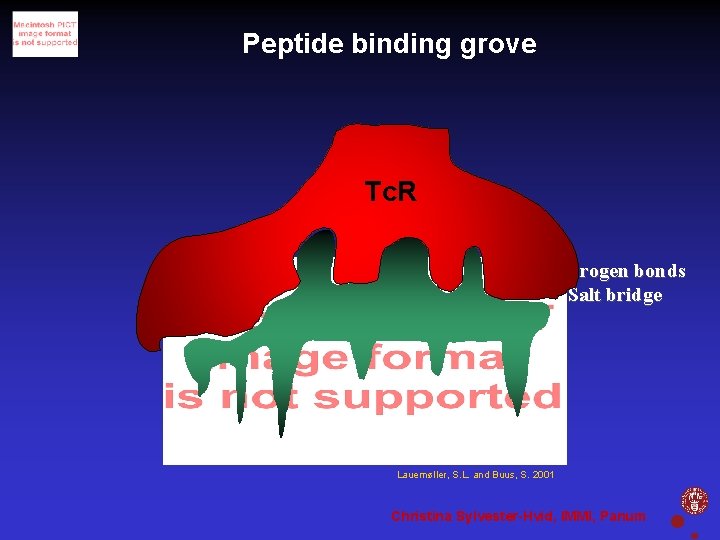
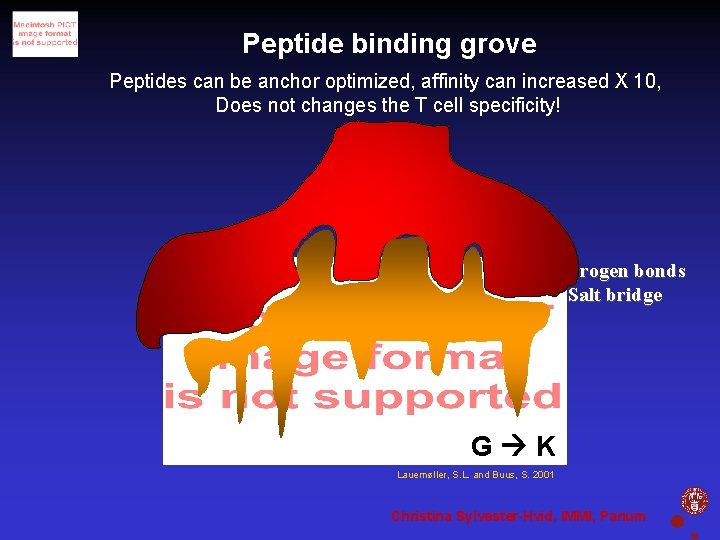
Peptide binding grove Tc. R Hydrogen bonds Salt bridge Hydrogen bonds Pockets Lauemøller, S. L. and Buus, S. 2001 Christina Sylvester-Hvid, IMMI, Panum

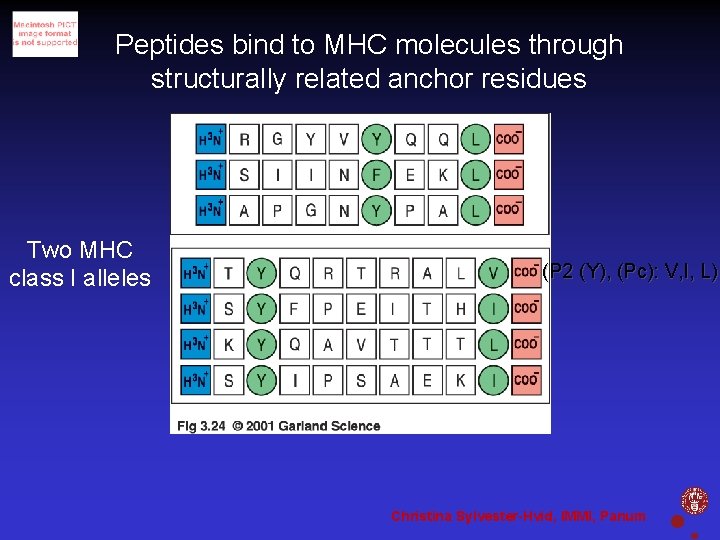
Peptides bind to MHC molecules through structurally related anchor residues Two MHC class I alleles (P 2 (Y), (Pc): V, I, L) Christina Sylvester-Hvid, IMMI, Panum

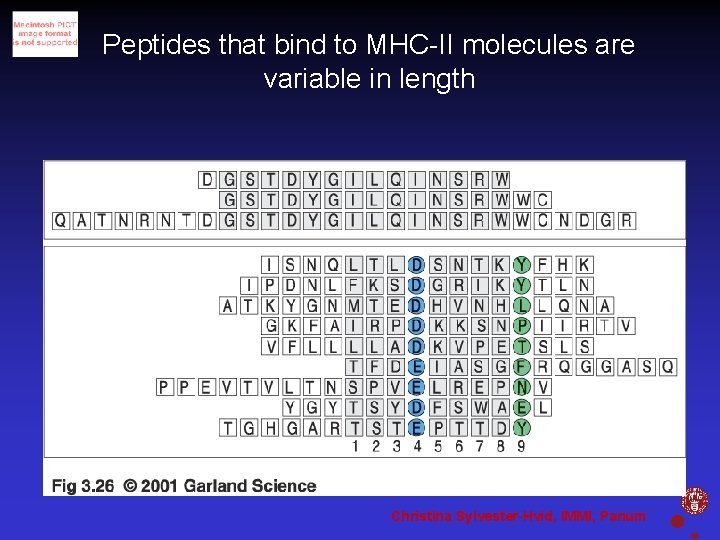
Peptides that bind to MHC-II molecules are variable in length Christina Sylvester-Hvid, IMMI, Panum

Comparison of the cleft architectures of murine class I alleles, Kb and Kk Stryhn A, et al. , 1996, PNAS Peptide: RGYVYQGL Peptide: FESTGNLI The peptide is deeply embedded Christina Sylvester-Hvid, IMMI, Panum

Most of the peptide is hidden in the groove - only a minor part is available for the Tc. R. FES T GN L I Available for the TCR Top view Mouse class I Kk in complex with peptide FESTGNLI Christina Sylvester-Hvid, IMMI, Panum

Peptide binding grove Peptides can be anchor optimized, affinity can increased X 10, Does not changes the T cell specificity! Hydrogen bonds Salt bridge Hydrogen bonds Pockets G K Lauemøller, S. L. and Buus, S. 2001 Christina Sylvester-Hvid, IMMI, Panum

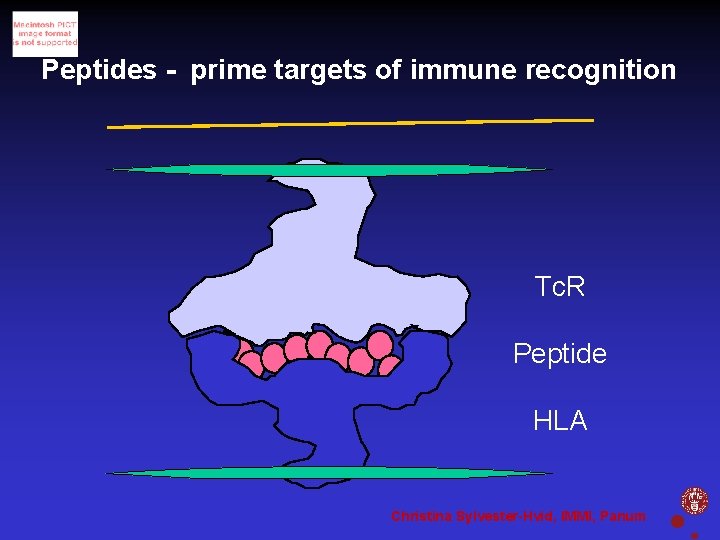
Peptides - prime targets of immune recognition Tc. R Peptide HLA Christina Sylvester-Hvid, IMMI, Panum

Determining primary protein structures is like charting the landscape of the immune system Christina Sylvester-Hvid, IMMI, Panum

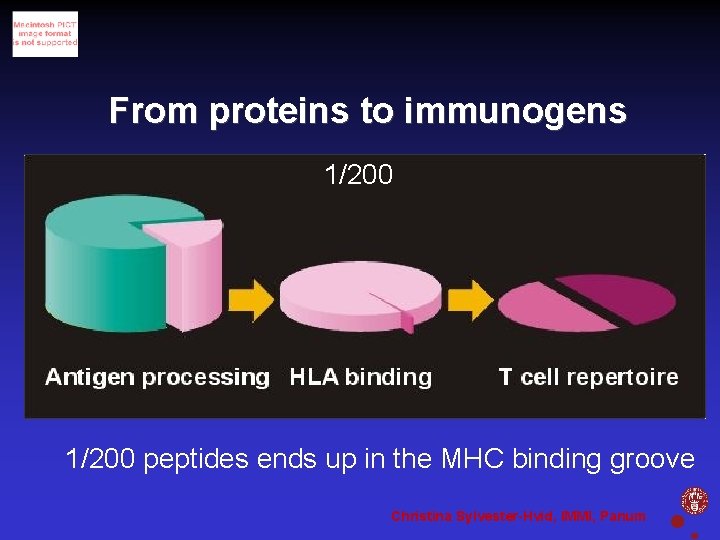
From proteins to immunogens 1/200 peptides ends up in the MHC binding groove Christina Sylvester-Hvid, IMMI, Panum

Translating genomes to immunogens Christina Sylvester-Hvid, IMMI, Panum

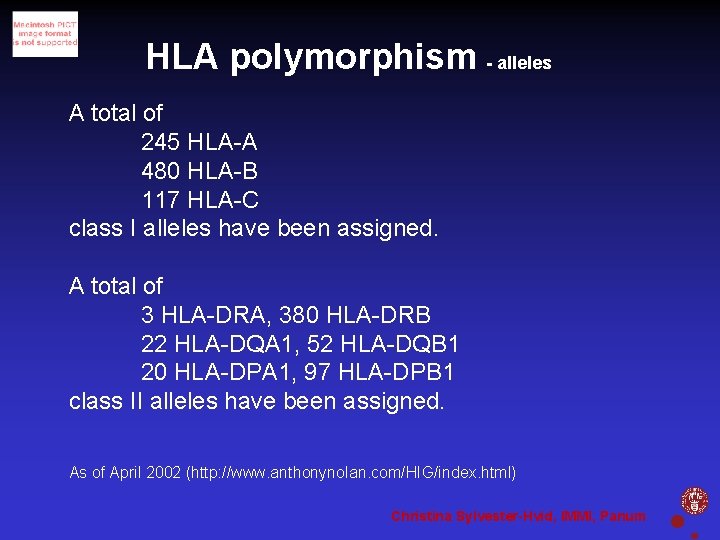
HLA polymorphism - alleles A total of 245 HLA-A 480 HLA-B 117 HLA-C class I alleles have been assigned. A total of 3 HLA-DRA, 380 HLA-DRB 22 HLA-DQA 1, 52 HLA-DQB 1 20 HLA-DPA 1, 97 HLA-DPB 1 class II alleles have been assigned. As of April 2002 (http: //www. anthonynolan. com/HIG/index. html) Christina Sylvester-Hvid, IMMI, Panum

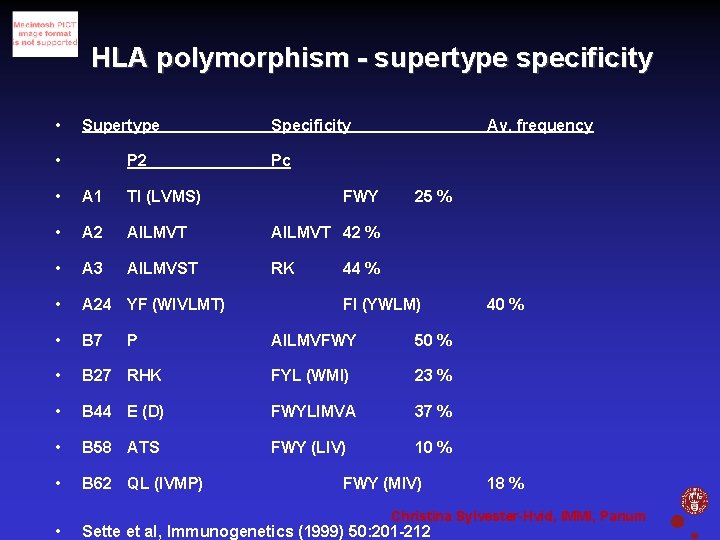
HLA polymorphism - supertype specificity • Supertype • P 2 Specificity Pc • A 1 TI (LVMS) • A 2 AILMVT 42 % • A 3 AILMVST RK • A 24 YF (WIVLMT) • B 7 • FWY 25 % 44 % FI (YWLM) AILMVFWY 50 % B 27 RHK FYL (WMI) 23 % • B 44 E (D) FWYLIMVA 37 % • B 58 ATS FWY (LIV) 10 % • B 62 QL (IVMP) • P Av. frequency FWY (MIV) 40 % 18 % Christina Sylvester-Hvid, IMMI, Panum Sette et al, Immunogenetics (1999) 50: 201 -212

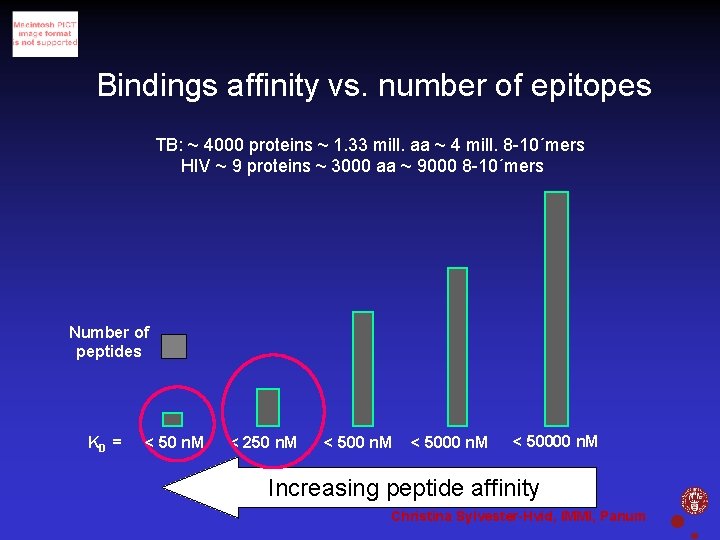
Bindings affinity vs. number of epitopes TB: ~ 4000 proteins ~ 1. 33 mill. aa ~ 4 mill. 8 -10´mers HIV ~ 9 proteins ~ 3000 aa ~ 9000 8 -10´mers Number of peptides KD = < 50 n. M < 250 n. M < 50000 n. M Increasing peptide affinity Christina Sylvester-Hvid, IMMI, Panum

Experimental description of peptide-MHC binding Many different peptide-MHC-binding assays have been suggested over the years ; without being exhaustive: 1) Olsen, A. C. , et al. , Eur. J. Immunol. 1994; 24: 385 -392 2) Buus, S et al. , J. Immunol. 1982; 129: 1883 -1891. 3 ) Buus, S. and Werdelin, O. J. Immunol. 1986; 136: 459 -465. 4) Babbitt, B. P et al. , Proc. Natl. Acad. Sci. USA. 1986; 83: 4509 -4513. 5) Buus, S. , et al. , Cell. 1986; 47: 1071 -1077. 6) Luescher, I. F et al. , Proc. Natl. Acad. Sci USA. 1988; 85: 871 -874. 7) Townsend, A. , et al. , Nature. 1989; 340: 443 -448. 8) Bouillot, M. et al. , Nature. 1989; 339: 473 -475. 9) Busch, R. and Rothbard, J. B. J. Immunol. Meth. 1990: 134: 1 -22. 10) Townsend, A. , et al. , Cell 1990: 62: 285 -295. 11) Parker, K. C. et al. , J. Immunol. 1992; 49: 1896 -1904. 12) Joosten, I. Et al. , Trans. Proc. 1993; 25: 2842 -2843. 13) Khilko, S. N. et al. , J. Biol. Chem. 1993; 268: 15425 -15434. 14) Regner, M. et al. , Exp Clin Immunogenet. 1996; 13: 30 -35. Christina Sylvester-Hvid, IMMI, Panum

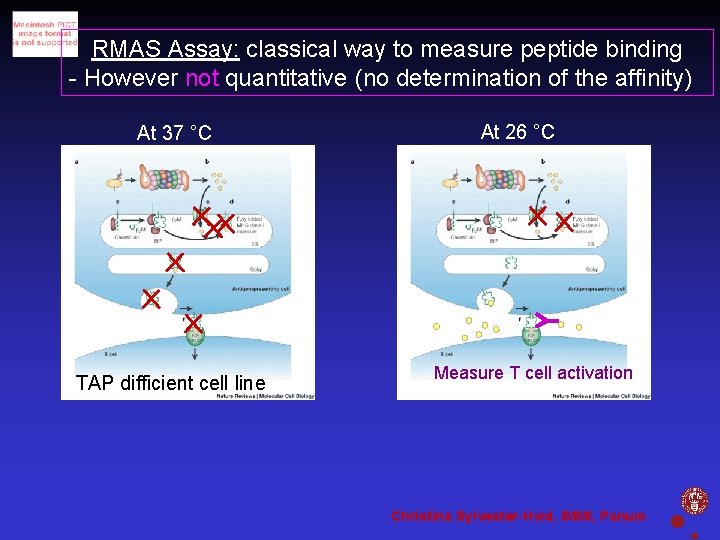
RMAS Assay: classical way to measure peptide binding - However not quantitative (no determination of the affinity) At 37 °C At 26 °C Add peptide TAP difficient cell line Measure T cell activation Christina Sylvester-Hvid, IMMI, Panum

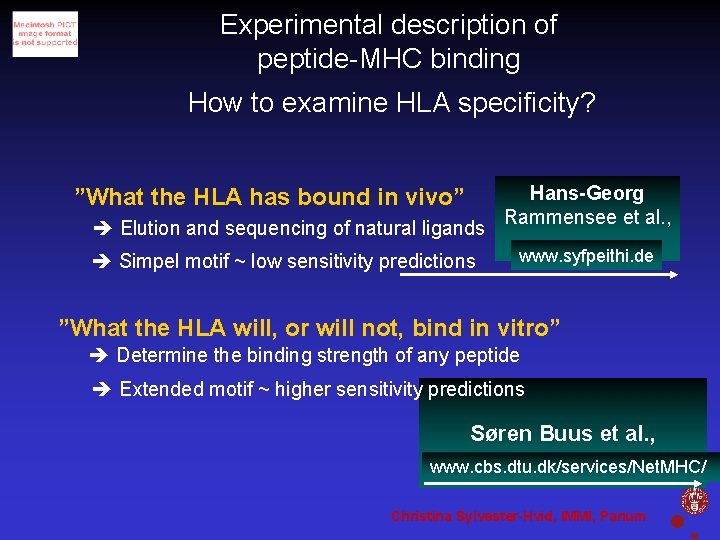
Experimental description of peptide-MHC binding How to examine HLA specificity? ”What the HLA has bound in vivo” Elution and sequencing of natural ligands Simpel motif ~ low sensitivity predictions Hans-Georg Rammensee et al. , www. syfpeithi. de ”What the HLA will, or will not, bind in vitro” Determine the binding strength of any peptide Extended motif ~ higher sensitivity predictions Søren Buus et al. , www. cbs. dtu. dk/services/Net. MHC/ Christina Sylvester-Hvid, IMMI, Panum

”What the HLA has bound in vivo” Prediction of binding, web based services (non quantitative) Christina Sylvester-Hvid, IMMI, Panum

www. syfpeithi. de (Hans-Georg Rammensee et al. , ) Christina Sylvester-Hvid, IMMI, Panum

� Søren Buus et al. , www. cbs. dtu. dk/services/Net. MHC/ Scanning the genome of Chlamydia pneumonia for CTL epitopes (n. M)
![How to determine peptide affinity Law of mass action koff R L kon How to determine peptide affinity Law of mass action koff [R] + [L] kon](https://slidetodoc.com/presentation_image/109932c5e1f17507c80f339796b20e83/image-25.jpg)
How to determine peptide affinity Law of mass action koff [R] + [L] kon koff [RL] [MHC] + [P] kon KD = koff (S-1)/kon (M-1 S-1) Binding hot peptide Saturation assay Binding 100% 50% Peptide [M] KD = (10 -15 -10 -6 M) [P*MHC] Peptide Log [M] Inhibition assay Cold Peptide Log [M] Log IC 50 Christina Sylvester-Hvid, IMMI, Panum

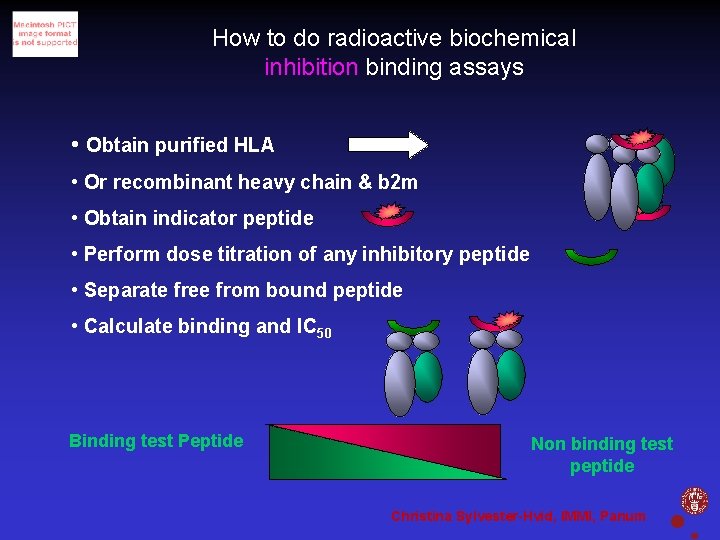
How to do radioactive biochemical inhibition binding assays • Obtain purified HLA • Or recombinant heavy chain & b 2 m • Obtain indicator peptide • Perform dose titration of any inhibitory peptide • Separate free from bound peptide • Calculate binding and IC 50 Binding test Peptide Non binding test peptide Christina Sylvester-Hvid, IMMI, Panum

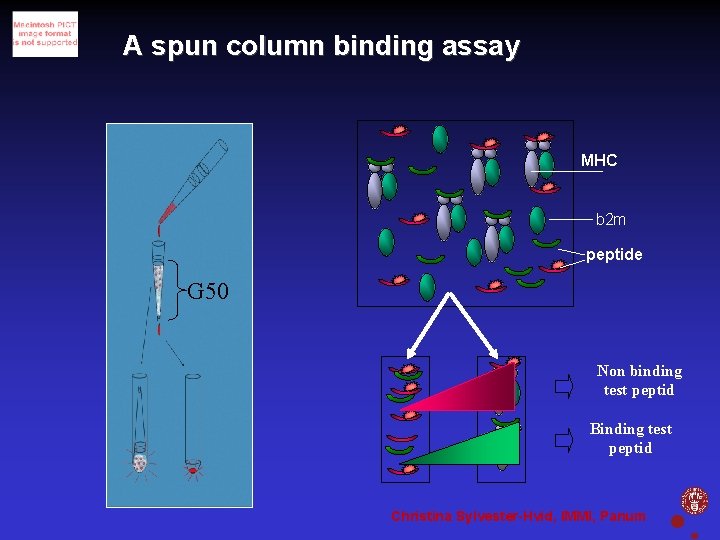
A spun column binding assay MHC b 2 m peptide G 50 Non binding test peptid Binding test peptid Christina Sylvester-Hvid, IMMI, Panum

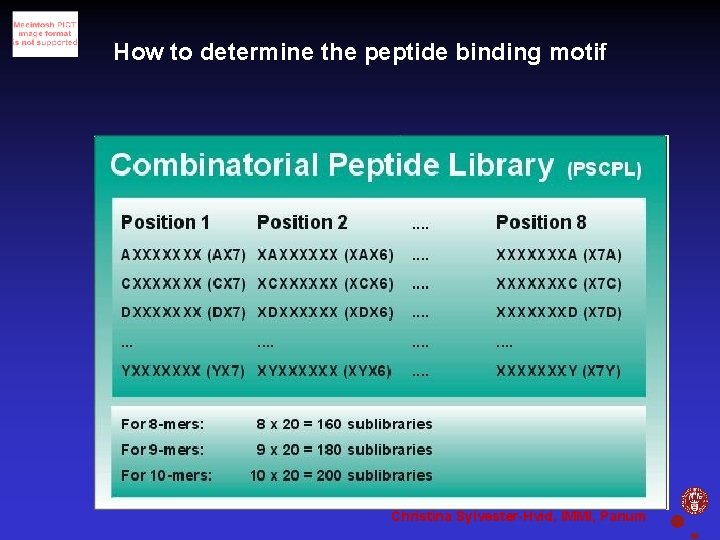
How to determine the peptide binding motif Christina Sylvester-Hvid, IMMI, Panum

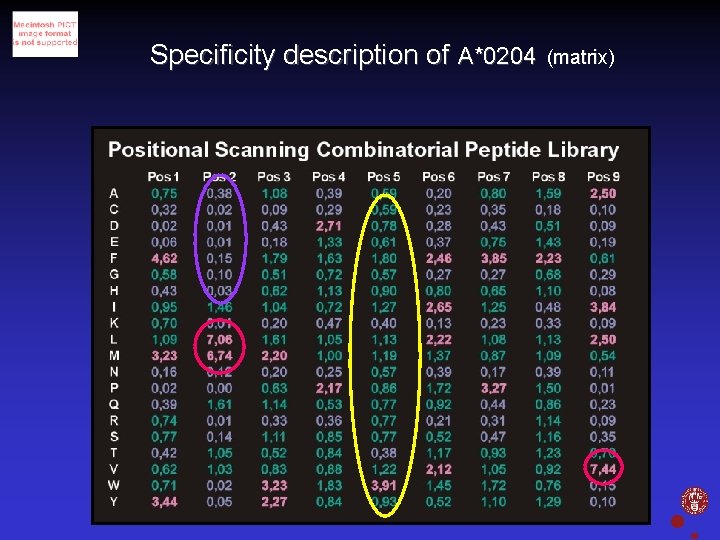
Specificity description of A*0204 (matrix) Christina Sylvester-Hvid, IMMI, Panum

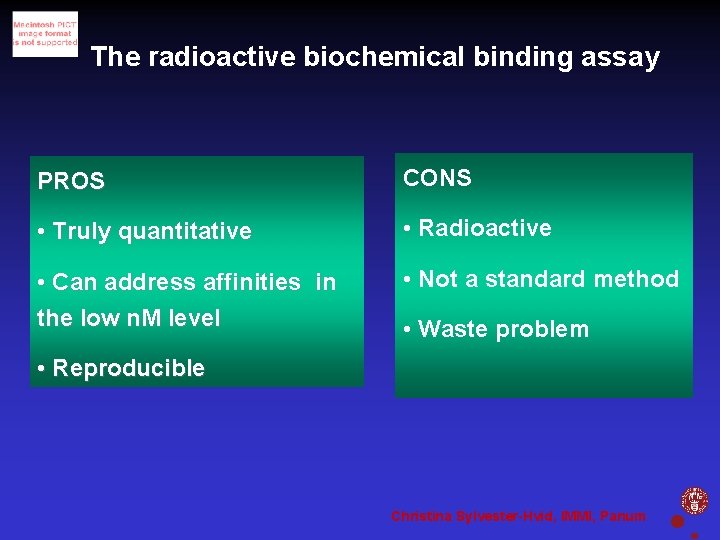
The radioactive biochemical binding assay PROS CONS • Truly quantitative • Radioactive • Can address affinities in the low n. M level • Not a standard method • Waste problem • Reproducible Christina Sylvester-Hvid, IMMI, Panum

The Quantitative ELISA Capable of Determining Peptide-MHC Class I Interaction • Made possible by our recent development of highly active recombinant MHC class I heavy chains – functional equivalents of ”empty” molecules L. O. Pedersen et al. , , EJI. 2001, 31: 2986 • Pros: • Reasonably simple, sensitive and quantitative • Does not depend on labeled peptide • It is easily adaptable to other laboratories • Disseminated protocol and standard reagents Christina Sylvester-Hvid, IMMI, Panum

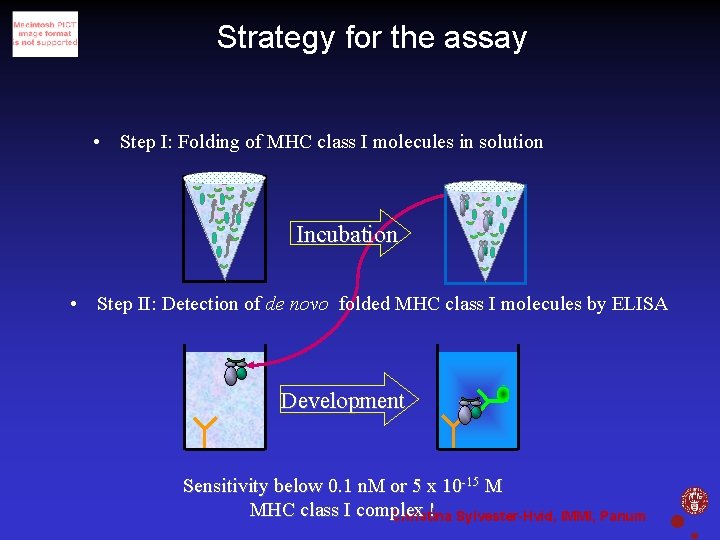
Strategy for the assay • Step I: Folding of MHC class I molecules in solution Incubation • Step II: Detection of de novo folded MHC class I molecules by ELISA Development Sensitivity below 0. 1 n. M or 5 x 10 -15 M MHC class I complex ! Sylvester-Hvid, IMMI, Panum Christina

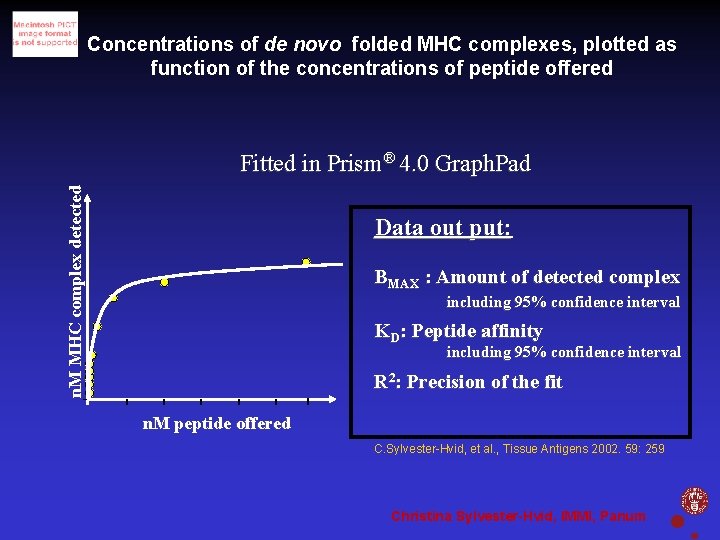
Concentrations of de novo folded MHC complexes, plotted as function of the concentrations of peptide offered n. M MHC complex detected Fitted in Prism® 4. 0 Graph. Pad Data out put: BMAX : Amount of detected complex including 95% confidence interval KD: Peptide affinity including 95% confidence interval R 2: Precision of the fit n. M peptide offered C. Sylvester-Hvid, et al. , Tissue Antigens 2002. 59: 259 Christina Sylvester-Hvid, IMMI, Panum

Data base of HLA ligands, founded by the NIH (Nat. Inst. Health) USA Christina Sylvester-Hvid, IMMI, Panum

Take home messages…. MHC class I molecule preferably binds peptides of 8 -11 aa Peptides bind to the MHC binding groove by hydrogen bonds, hydrophobic forces and other non-covalent interactions MHC binding specificity is obtained through the recognition of peptide- motifs, a recognition mode which requires the presence and proper spacing of particular amino acids in certain anchor positions. The binding strength, the affinity - is very important: The higher affinity of a peptide to the class I molecule, the higher chance of being immunogenic. Christina Sylvester-Hvid, IMMI, Panum

and more…. . http: //www. ihwg. org/components/peptider. htm Peptide binding can be addressed in a qualitative or quantitative matter. The Radioactive binding assay or The quantitative ELISA assay can measure the exact affinity in n. M for the best known binders Measurements of affinity can be used to generate tools (ANN) for prediction of peptide binding to any MHC class I molecule of human interest. Subsequently to validation, peptides can be directly used in peptide based vaccines or rationally optimized to increase their immunogenesity Christina Sylvester-Hvid, IMMI, Panum