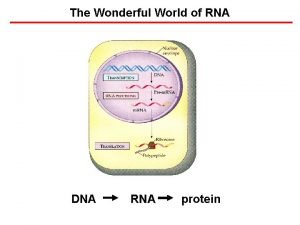
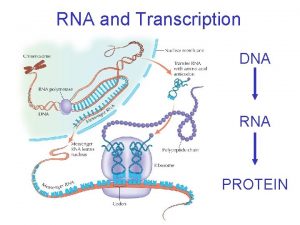
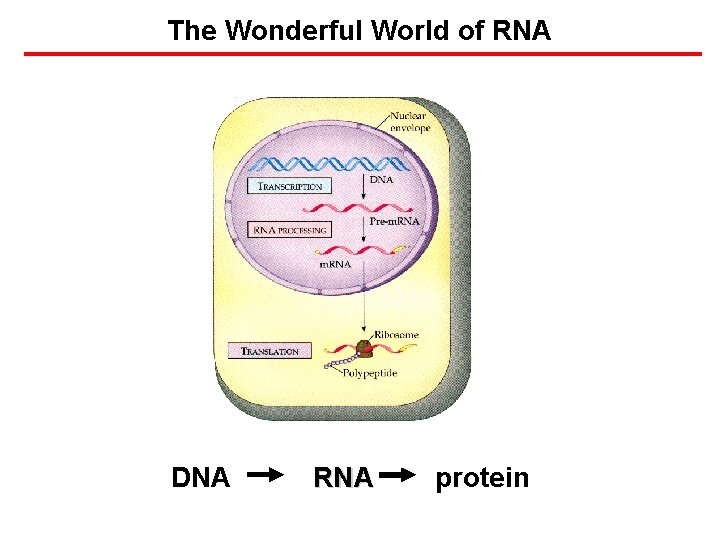
The Wonderful World of RNA DNA RNA protein


- Slides: 15

The Wonderful World of RNA DNA RNA protein

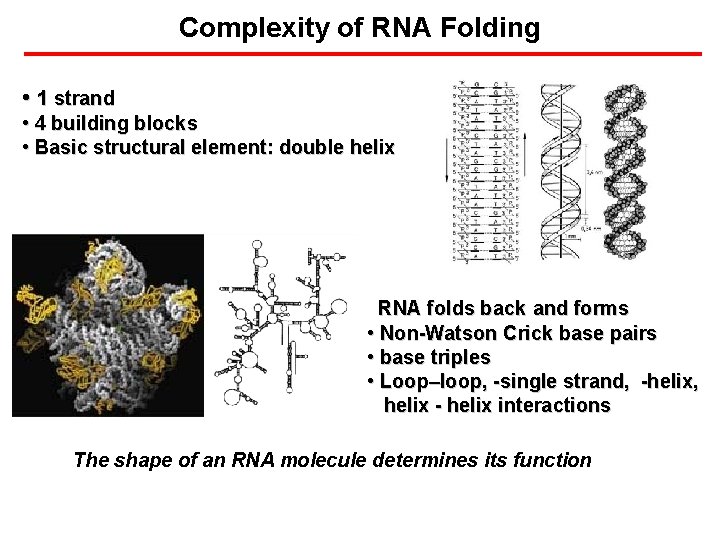
Complexity of RNA Folding • 1 strand • 4 building blocks • Basic structural element: double helix RNA folds back and forms • Non-Watson Crick base pairs • base triples • Loop–loop, -single strand, -helix, helix - helix interactions The shape of an RNA molecule determines its function

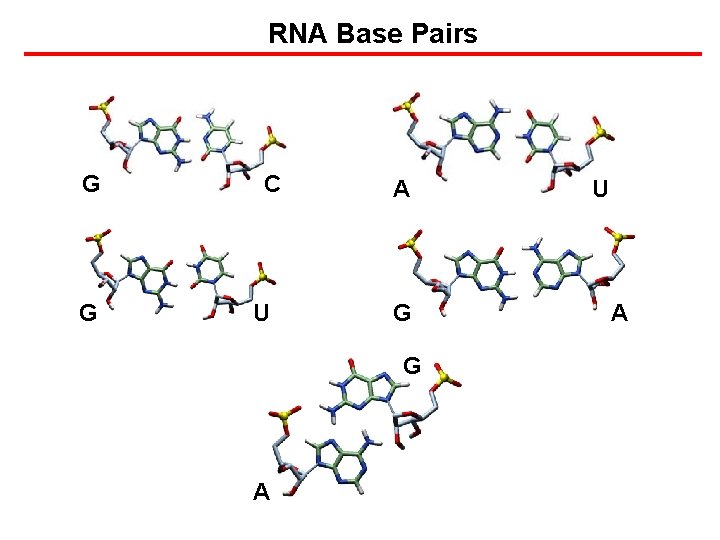
RNA Base Pairs G G C U A G G A U A

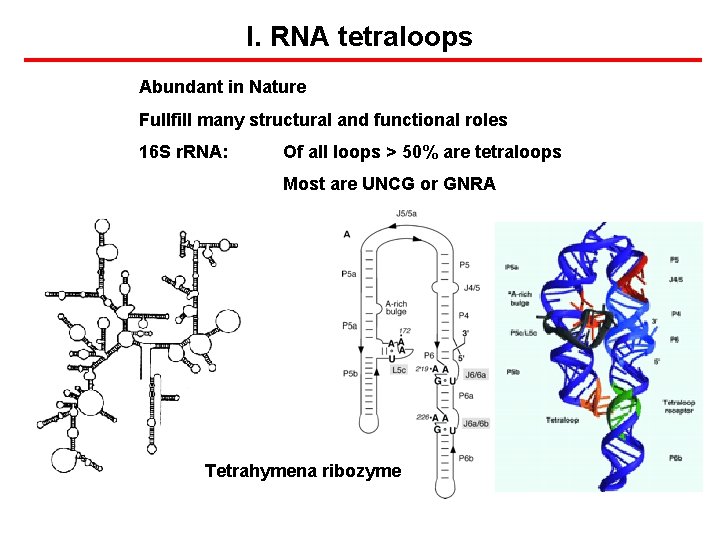
I. RNA tetraloops Abundant in Nature Fullfill many structural and functional roles 16 S r. RNA: Of all loops > 50% are tetraloops Most are UNCG or GNRA Tetrahymena ribozyme

II. Cis-regulating elements of poliovirus Single stranded RNA genome 7440 nt

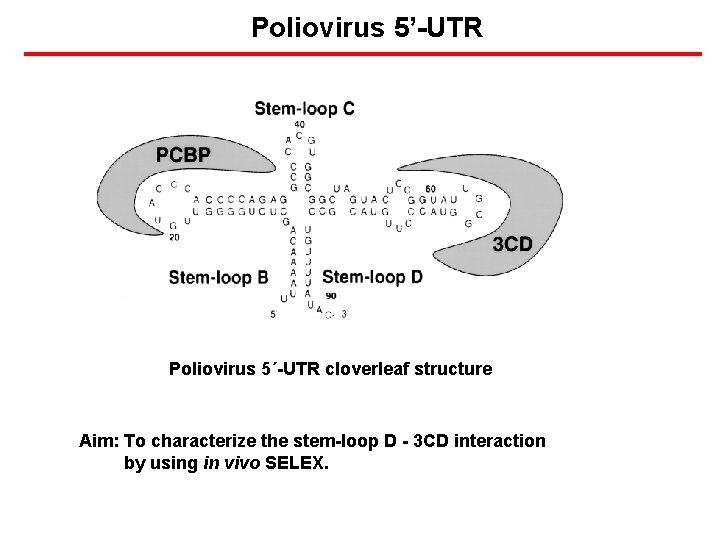
Poliovirus 5’-UTR Poliovirus 5´-UTR cloverleaf structure Aim: To characterize the stem-loop D - 3 CD interaction by using in vivo SELEX.

In vivo selection of randomly generated mutants polio wild type sequence N N N-N U-A G-C random pool of 48 = 65536 possible sequences GC U G U-G A-U U-A G-C In vivo production of virus mutants N N N-N U-A G-C 22 different selected sequences

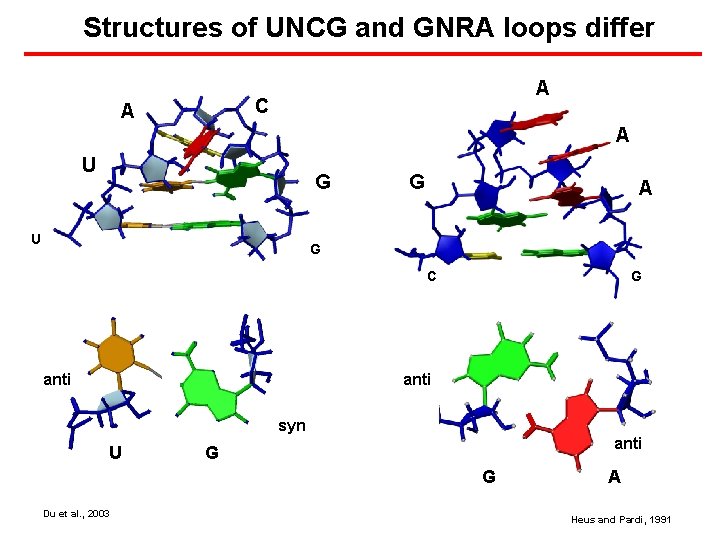
Structures of UNCG and GNRA loops differ A C A A U G A G C anti G anti syn U anti G G Du et al. , 2003 A Heus and Pardi, 1991

II. RNA pseudoknots • Occur in virtually all classes of RNA • Role in biological processes: - replication initiation - translational control - ribosomal frameshifting - core formation in larger RNAs

RNA pseudoknot formation

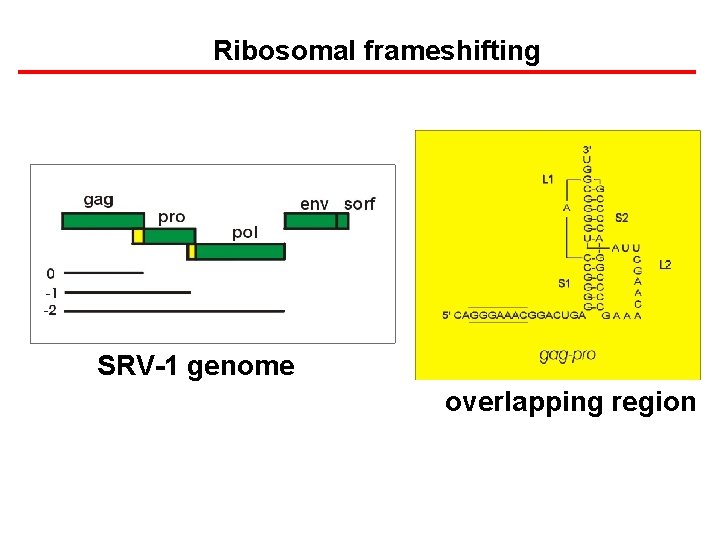
Ribosomal frameshifting SRV-1 genome overlapping region

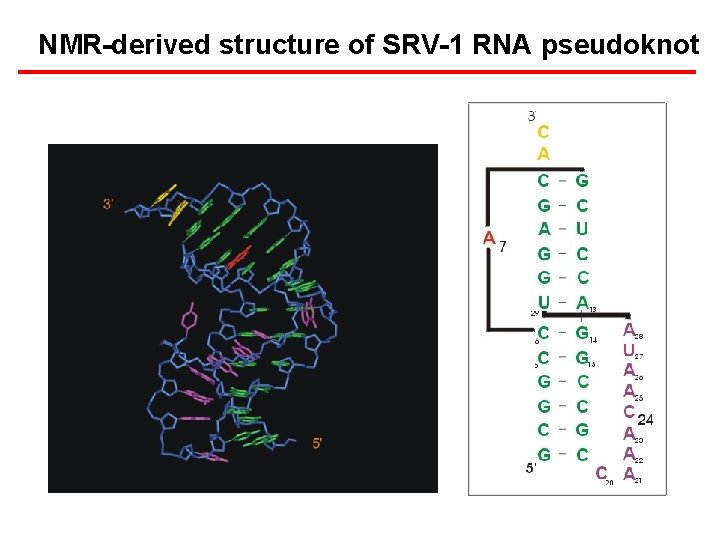
NMR-derived structure of SRV-1 RNA pseudoknot

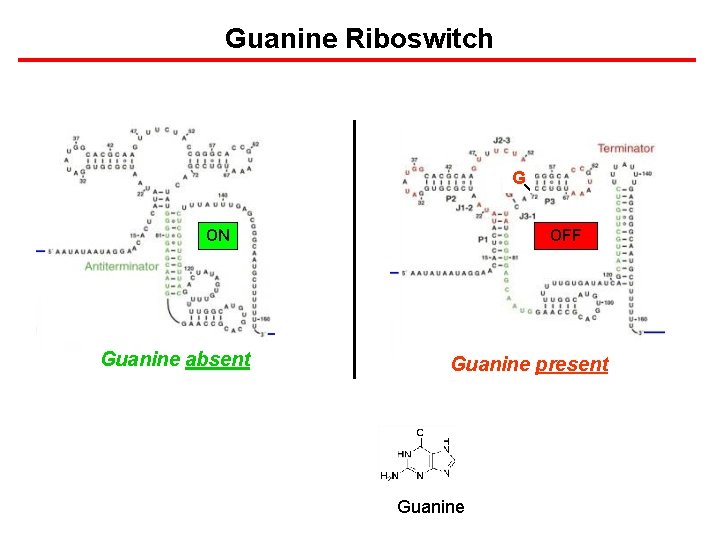
Guanine Riboswitch G ON Guanine absent OFF Guanine present Guanine

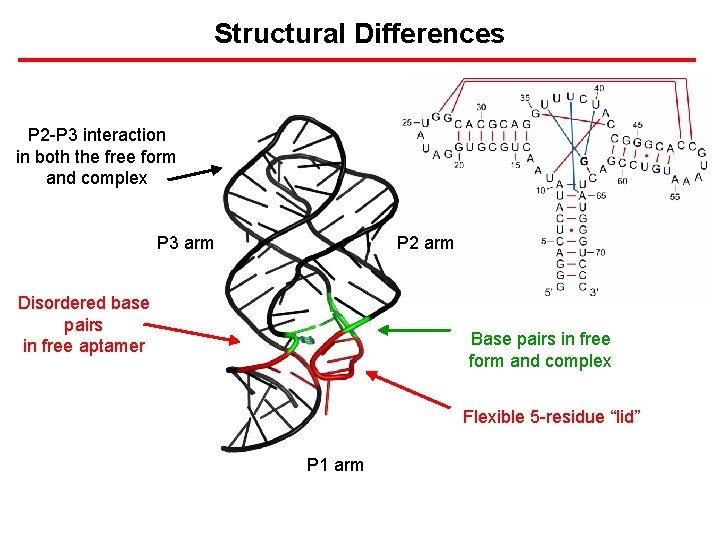
Structural Differences P 2 -P 3 interaction in both the free form and complex P 3 arm P 2 arm Disordered base pairs in free aptamer Base pairs in free form and complex Flexible 5 -residue “lid” P 1 arm

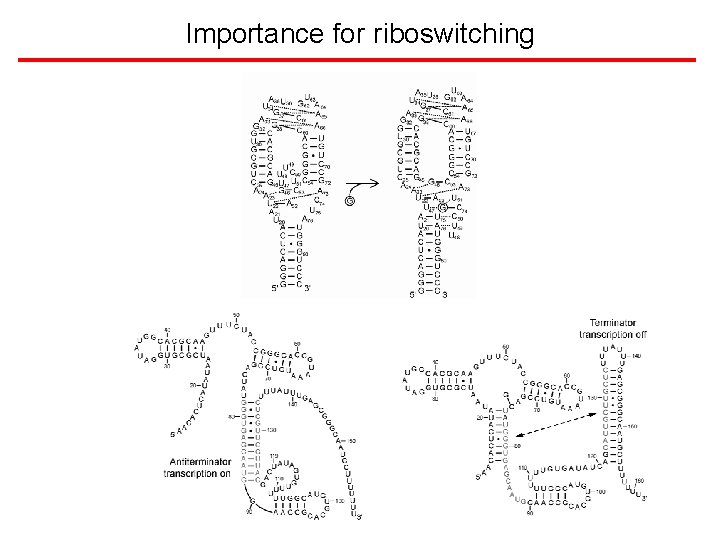
Importance for riboswitching
 Dna and genes chapter 11
Dna and genes chapter 11 Dna to protein steps
Dna to protein steps Mutations quiz
Mutations quiz Chapter 12 section 3 dna rna and protein
Chapter 12 section 3 dna rna and protein Dna protein synthesis study guide answers
Dna protein synthesis study guide answers Microarray analysis
Microarray analysis Dna rna protein
Dna rna protein Dna rna protein diagram
Dna rna protein diagram Dna rna protein trait
Dna rna protein trait Wonderful so wonderful is your unfailing love
Wonderful so wonderful is your unfailing love Wonderful wonderful jesus
Wonderful wonderful jesus Here i go out to sea again
Here i go out to sea again Rna protein synthesis
Rna protein synthesis Section 12-3 rna and protein synthesis answer key
Section 12-3 rna and protein synthesis answer key Order of bases in dna
Order of bases in dna Rna transfer
Rna transfer