The use of 4 grams for Protein Classification





- Slides: 21

The use of 4 -grams for Protein Classification and Sequence Comparison Dror Tobi, Shann. Ching Chen, Ivet Bahar

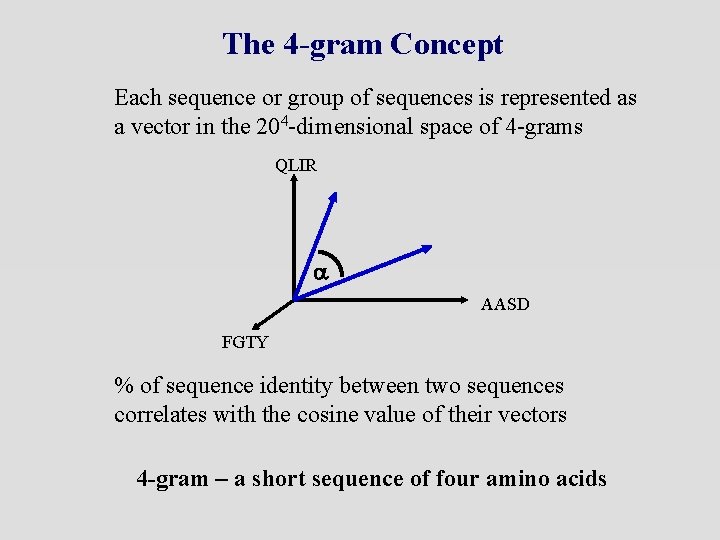
The 4 -gram Concept Each sequence or group of sequences is represented as a vector in the 204 -dimensional space of 4 -grams QLIR a AASD FGTY % of sequence identity between two sequences correlates with the cosine value of their vectors 4 -gram – a short sequence of four amino acids

Representation of Sequence(s) as 4 -gram Vector(s) Three steps: Ø Calculating 4 -gram frequencies in the examined DB Ø Calculating 4 -gram frequencies for a given sequence or a given family of sequences Ø Creating a 4 -gram vector using a weight function

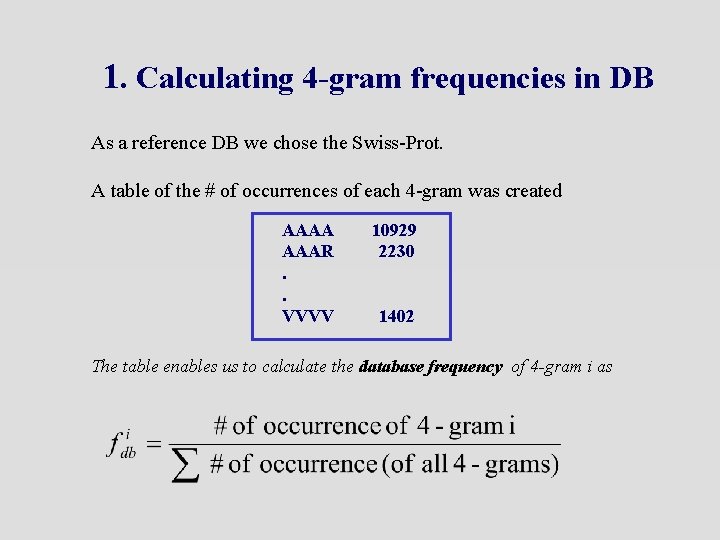
1. Calculating 4 -gram frequencies in DB As a reference DB we chose the Swiss-Prot. A table of the # of occurrences of each 4 -gram was created AAAA AAAR. . VVVV 10929 2230 1402 The table enables us to calculate the database frequency of 4 -gram i as

2. Calculating 4 -gram frequencies of a sequence (or family) The 4 -gram frequencies for a given sequence or a family of sequences is done using a hash table. Each 4 -gram is entered into a hash table from which the 4 gram family frequency is calculated xxxx n

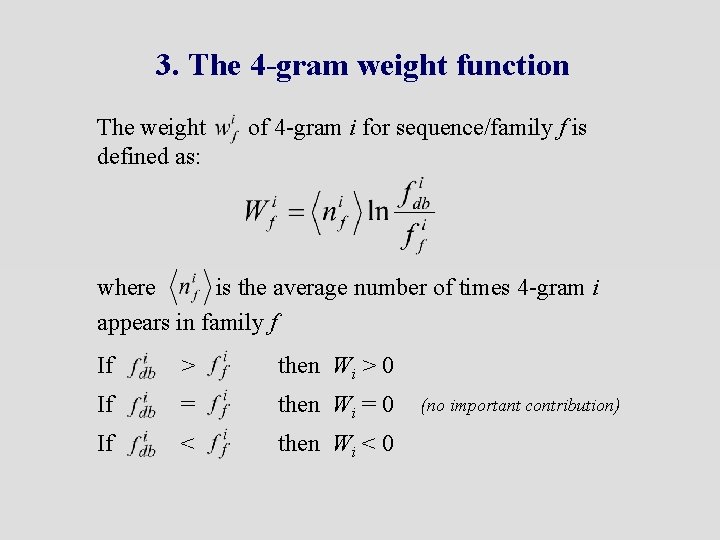
3. The 4 -gram weight function The weight defined as: of 4 -gram i for sequence/family f is where is the average number of times 4 -gram i appears in family f If > then Wi > 0 If = then Wi = 0 If < then Wi < 0 (no important contribution)

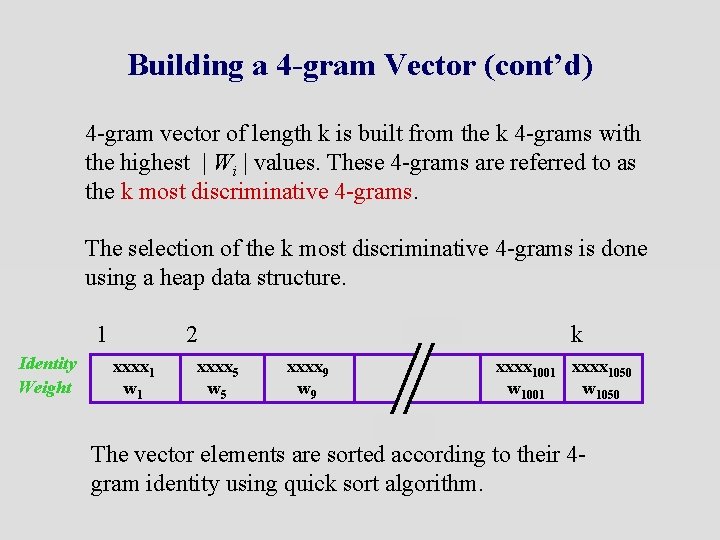
Building a 4 -gram Vector (cont’d) 4 -gram vector of length k is built from the k 4 -grams with the highest | Wi | values. These 4 -grams are referred to as the k most discriminative 4 -grams. The selection of the k most discriminative 4 -grams is done using a heap data structure. 1 Identity Weight 2 xxxx 1 w 1 xxxx 5 w 5 k xxxx 9 w 9 xxxx 1001 xxxx 1050 w 1001 w 1050 The vector elements are sorted according to their 4 gram identity using quick sort algorithm.

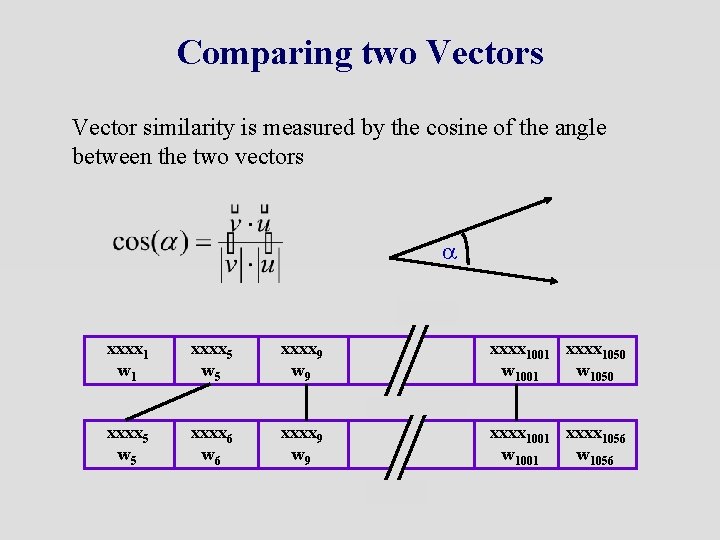
Comparing two Vectors Vector similarity is measured by the cosine of the angle between the two vectors a xxxx 1 w 1 xxxx 5 w 5 xxxx 9 w 9 xxxx 1001 xxxx 1050 w 1001 w 1050 xxxx 5 w 5 xxxx 6 w 6 xxxx 9 w 9 xxxx 1001 xxxx 1056 w 1001 w 1056

EC 4 family classification EC 4 Test 1769 families (containing a total of 10, 919 enzymes) defined at the EC level 4 classification (at Expasy) were considered (*). A 4 -gram vector (model, probe vector) was built for each EC 4 family. The cosine between the probe vector for a given EC 4 family and the 4 -gram vector of each sequence in the Swiss-Prot was calculated. All sequences were rankordered based on their cosine values. (*) out of a total of ~4000 in SWISS-PROT release 27. 7, excluding families that do not contain any sequences

Success Definition % success is defined as the % of family members having a cosine value higher then any non family sequence in the Swiss-Prot DB. Example: for a family (F 00 X) that has five members F 001 -5 A case of 80% success. Family members are colored blue. F 001 F 003 F 005 F 002 P 0 SD F 004 …. . 0. 567 0. 456 0. 354 0. 333 0. 301 0. 255

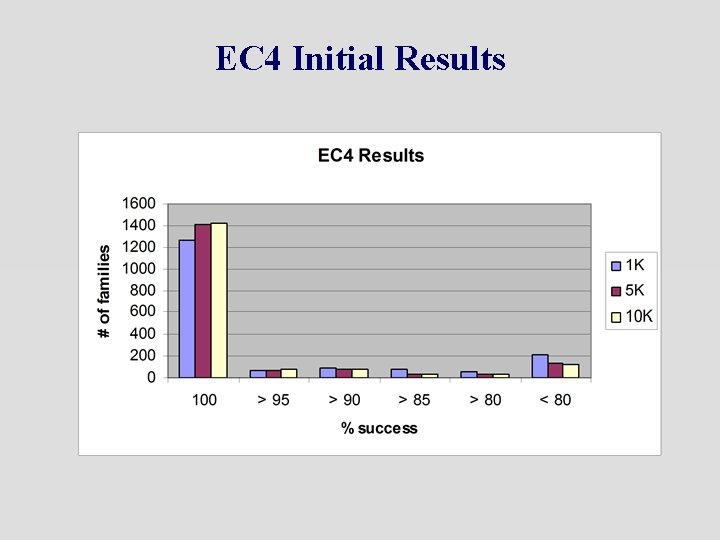
EC 4 Initial Results

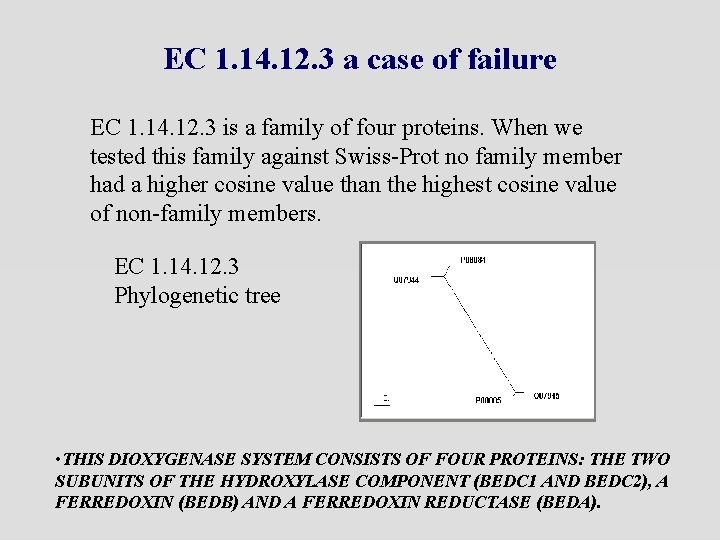
EC 1. 14. 12. 3 a case of failure EC 1. 14. 12. 3 is a family of four proteins. When we tested this family against Swiss-Prot no family member had a higher cosine value than the highest cosine value of non-family members. EC 1. 14. 12. 3 Phylogenetic tree • THIS DIOXYGENASE SYSTEM CONSISTS OF FOUR PROTEINS: THE TWO SUBUNITS OF THE HYDROXYLASE COMPONENT (BEDC 1 AND BEDC 2), A FERREDOXIN (BEDB) AND A FERREDOXIN REDUCTASE (BEDA).

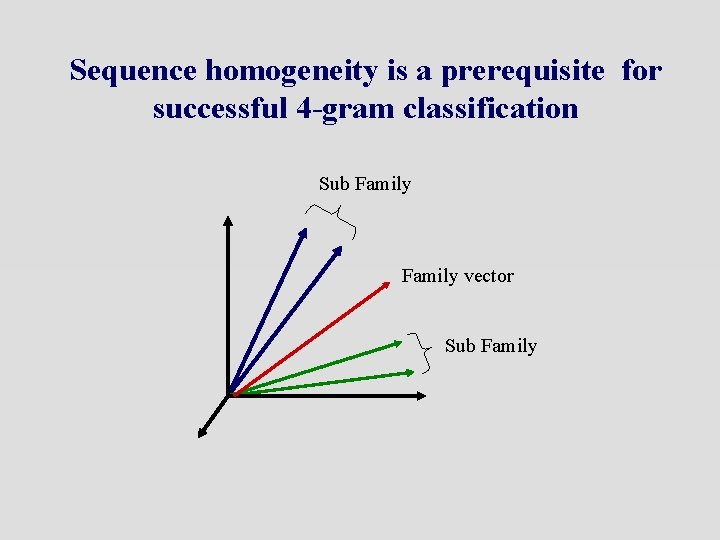
Sequence homogeneity is a prerequisite for successful 4 -gram classification Sub Family vector Sub Family

Preliminary Conclusions Ø 4 -gram classification is a fast way to classify/cluster sequences. 120, 000 comparisons take ~4 min on regular desktop. ØSequence homogeneity within a family is a prerequisite for successful classification. ØThe EC classification classifies enzymes according to their function, which does not necessarily correlate with classification based upon sequence similarity.

4 -grams uses in Sequence Search The 4 -gram vector “as is” measures “sequence identity” and therefore can easily detect close sequences ( >55% identity) But what about sequences with low sequence identity (30 -55%)?

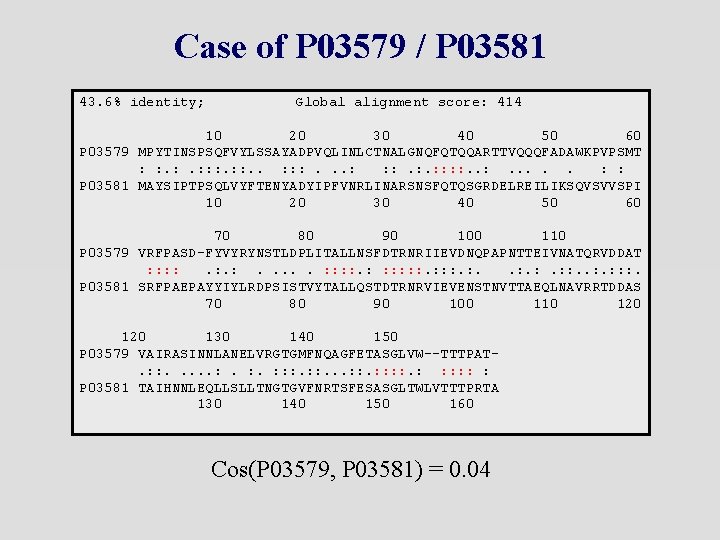
Case of P 03579 / P 03581 43. 6% identity; Global alignment score: 414 10 20 30 40 50 60 P 03579 MPYTINSPSQFVYLSSAYADPVQLINLCTNALGNQFQTQQARTTVQQQFADAWKPVPSMT : : : . . . : : P 03581 MAYSIPTPSQLVYFTENYADYIPFVNRLINARSNSFQTQSGRDELREILIKSQVSVVSPI 10 20 30 40 50 60 70 80 90 100 110 P 03579 VRFPASD-FYVYRYNSTLDPLITALLNSFDTRNRIIEVDNQPAPNTTEIVNATQRVDDAT : : . : . . . : : : . : . : : : . P 03581 SRFPAEPAYYIYLRDPSISTVYTALLQSTDTRNRVIEVENSTNVTTAEQLNAVRRTDDAS 70 80 90 100 110 120 130 140 150 P 03579 VAIRASINNLANELVRGTGMFNQAGFETASGLVW--TTTPAT. : : . . . : . : : : . . . : : : : P 03581 TAIHNNLEQLLSLLTNGTGVFNRTSFESASGLTWLVTTTPRTA 130 140 150 160 Cos(P 03579, P 03581) = 0. 04

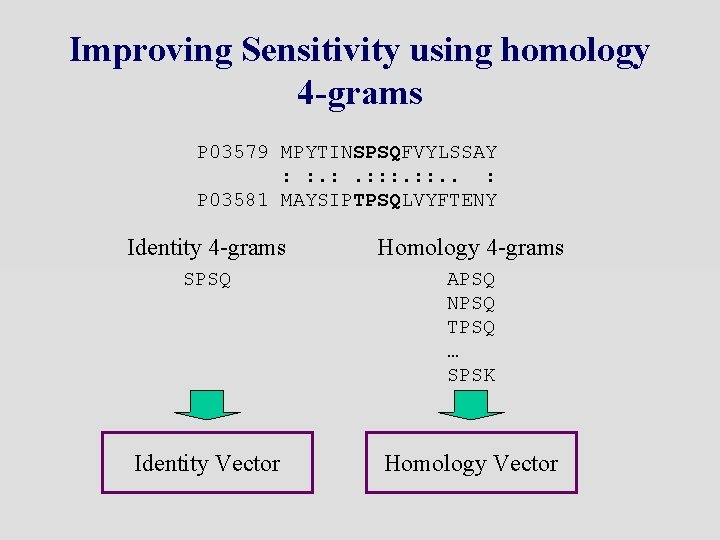
Improving Sensitivity using homology 4 -grams P 03579 MPYTINSPSQFVYLSSAY : : : . . : P 03581 MAYSIPTPSQLVYFTENY Identity 4 -grams Homology 4 -grams SPSQ APSQ NPSQ TPSQ … SPSK Identity Vector Homology Vector

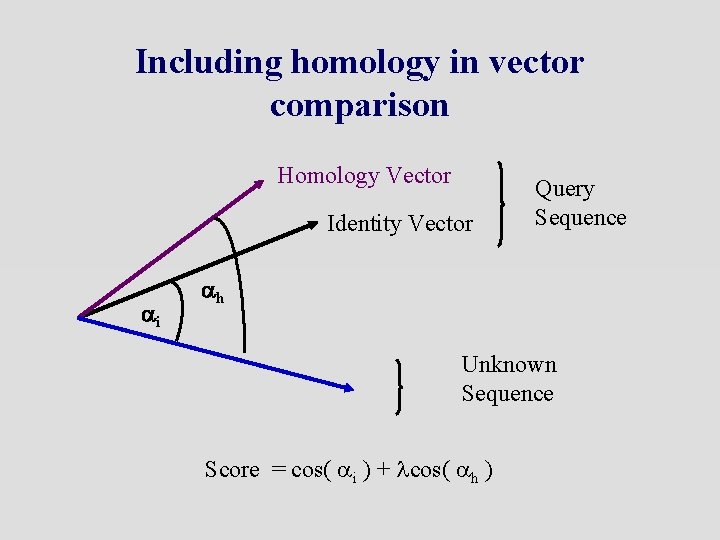
Including homology in vector comparison Homology Vector Identity Vector ai Query Sequence ah Unknown Sequence Score = cos( ai ) + lcos( ah )

4 -gram Search Results

Correlation between cosine value and Sequence alignment % identity

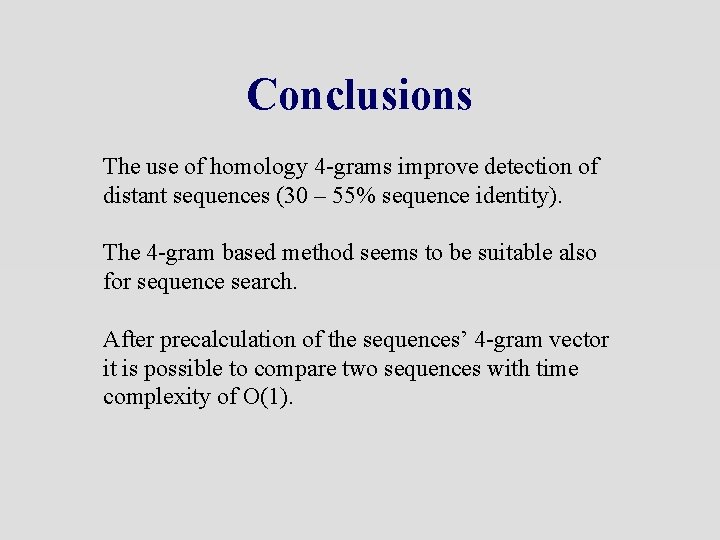
Conclusions The use of homology 4 -grams improve detection of distant sequences (30 – 55% sequence identity). The 4 -gram based method seems to be suitable also for sequence search. After precalculation of the sequences’ 4 -gram vector it is possible to compare two sequences with time complexity of O(1).
 Protein pump vs protein channel
Protein pump vs protein channel Protein-protein docking
Protein-protein docking Benjamin classification
Benjamin classification Functional classification of protein
Functional classification of protein Protein definition and classification
Protein definition and classification Protein classification based on composition
Protein classification based on composition Kontinuitetshantering
Kontinuitetshantering Typiska novell drag
Typiska novell drag Tack för att ni lyssnade bild
Tack för att ni lyssnade bild Ekologiskt fotavtryck
Ekologiskt fotavtryck Varför kallas perioden 1918-1939 för mellankrigstiden?
Varför kallas perioden 1918-1939 för mellankrigstiden? En lathund för arbete med kontinuitetshantering
En lathund för arbete med kontinuitetshantering Kassaregister ideell förening
Kassaregister ideell förening Tidböcker
Tidböcker Anatomi organ reproduksi
Anatomi organ reproduksi Densitet vatten
Densitet vatten Datorkunskap för nybörjare
Datorkunskap för nybörjare Boverket ka
Boverket ka Hur skriver man en debattartikel
Hur skriver man en debattartikel Autokratiskt ledarskap
Autokratiskt ledarskap Nyckelkompetenser för livslångt lärande
Nyckelkompetenser för livslångt lärande Påbyggnader för flakfordon
Påbyggnader för flakfordon