The Steroid and Xenobiotic Receptor SXR A Key

The Steroid and Xenobiotic Receptor SXR: A Key Regulator of Drug and Xenobiotic Metabolism Bruce Blumberg, Ph. D. Department of Developmental and Cell Biology Center for Biomedical Engineering Institute for Genomics and Bioinformatics University of California, Irvine
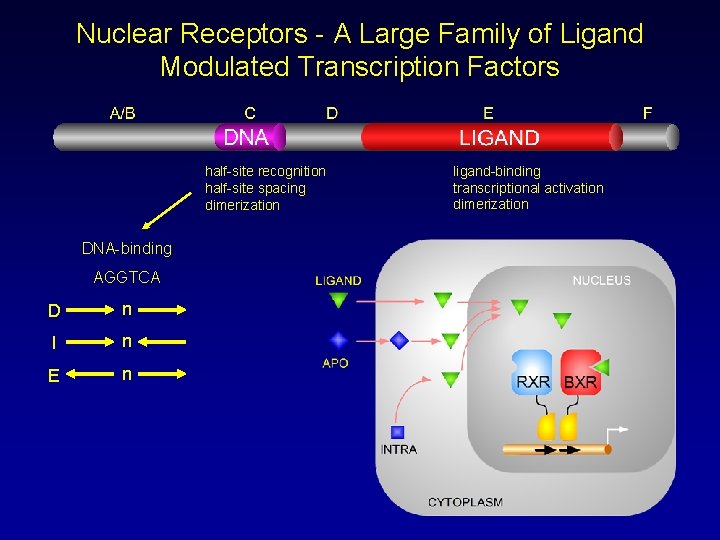
Nuclear Receptors - A Large Family of Ligand Modulated Transcription Factors half-site recognition half-site spacing dimerization DNA-binding AGGTCA D n I n E n ligand-binding transcriptional activation dimerization
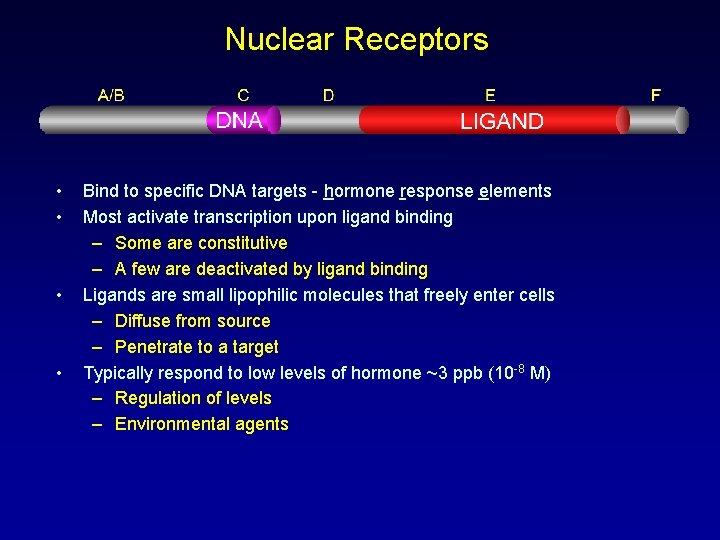
Nuclear Receptors • • Bind to specific DNA targets - hormone response elements Most activate transcription upon ligand binding – Some are constitutive – A few are deactivated by ligand binding Ligands are small lipophilic molecules that freely enter cells – Diffuse from source – Penetrate to a target Typically respond to low levels of hormone ~3 ppb (10 -8 M) – Regulation of levels – Environmental agents

Mithridates VI Eupator The Royal Toxicologist (120 -63 BC) King of Pontus aka Mithridates the Great

Long Standing Questions • Mithridatum - generalized tolerance to poison • Adaptive hepatic response (Hans Selye) – Exposure to certain “catatoxic” chemicals elicits protection against later exposure to others – Apparently mediated via CYP upregulation • What is the mechanism?
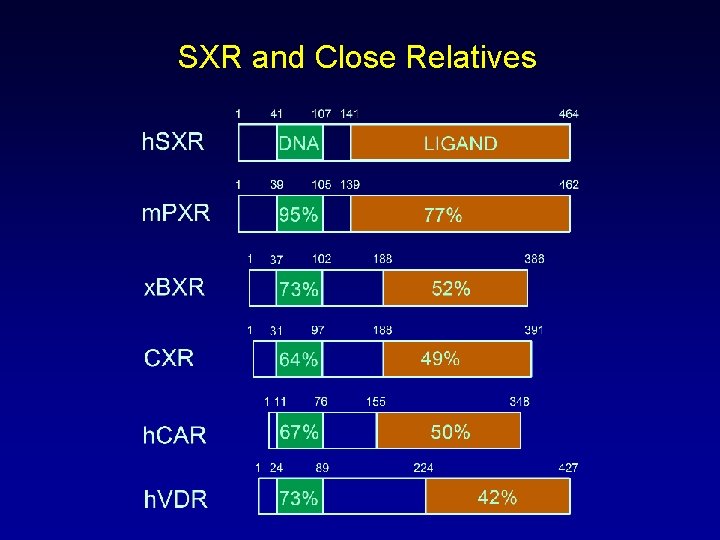
SXR and Close Relatives
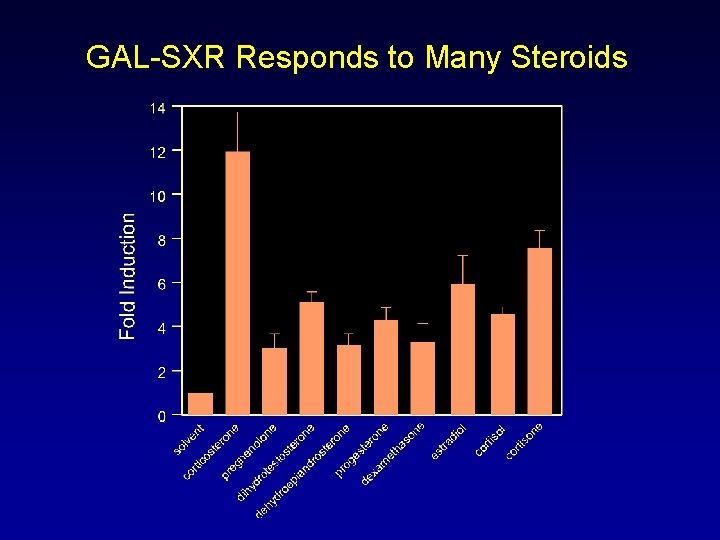
GAL-SXR Responds to Many Steroids
The Steroid Sensor Hypothesis • Removal of bioactive steroids and xenobiotics is required for physiologic homeostasis • Steroid production is regulated, why not catabolism? • Hundreds of steroid metabolites make it unreasonable to have individual regulation • Hypothesize a broad specificity sensor that monitors steroid levels and regulates the expression of degradative enzymes, e. G. P 450 s – Broad specificity probably necessitates low-affinity
Predictions and Requirements of the Model • Sensor should be expressed in tissues that catabolize steroids and xenobiotics • Catabolic enzymes should be targets for the sensor • Compounds known to induce catabolic enzymes should activate the sensor • Partially metabolized (reduced) steroids should activate sensor
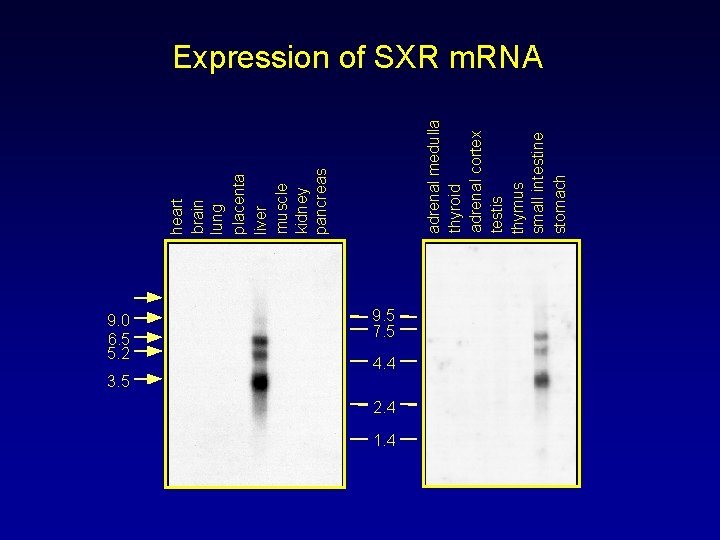
9. 0 6. 5 5. 2 3. 5 adrenal medulla thyroid adrenal cortex testis thymus small intestine stomach heart brain lung placenta liver muscle kidney pancreas Expression of SXR m. RNA 9. 5 7. 5 4. 4 2. 4 1. 4
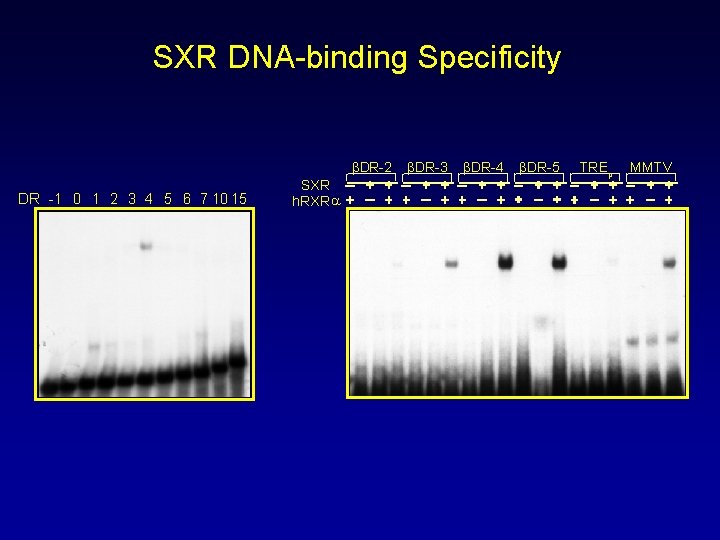
SXR DNA-binding Specificity βDR-2 DR -1 0 1 2 3 4 5 6 7 10 15 SXR h. RXR a βDR-3 βDR-4 βDR-5 TREp MMTV
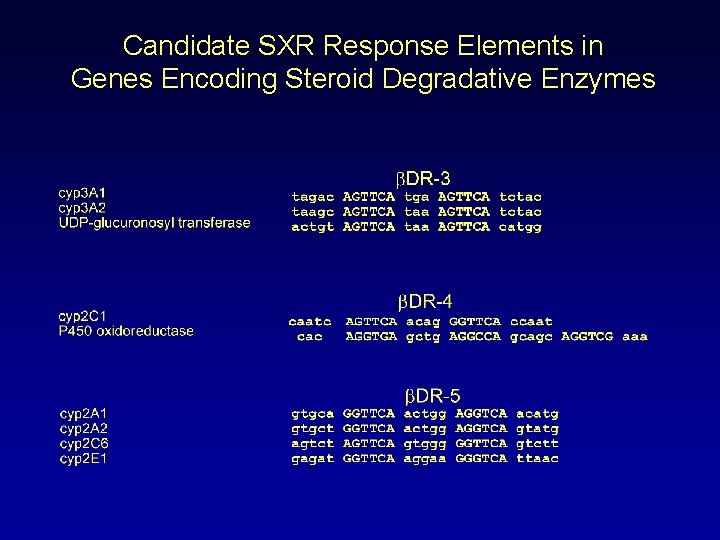
Candidate SXR Response Elements in Genes Encoding Steroid Degradative Enzymes
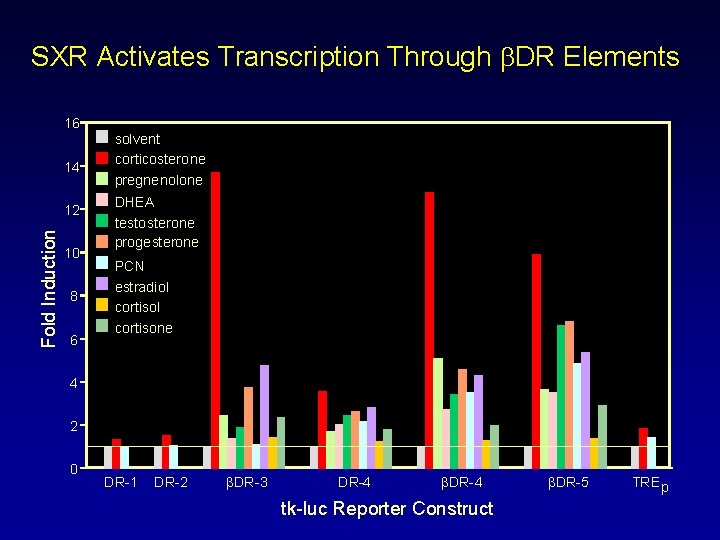
SXR Activates Transcription Through b. DR Elements 16 14 Fold Induction 12 10 8 6 solvent corticosterone pregnenolone DHEA testosterone progesterone PCN estradiol cortisone 4 2 0 DR-1 DR-2 βDR-3 DR-4 βDR-4 tk-luc Reporter Construct βDR-5 TRE p
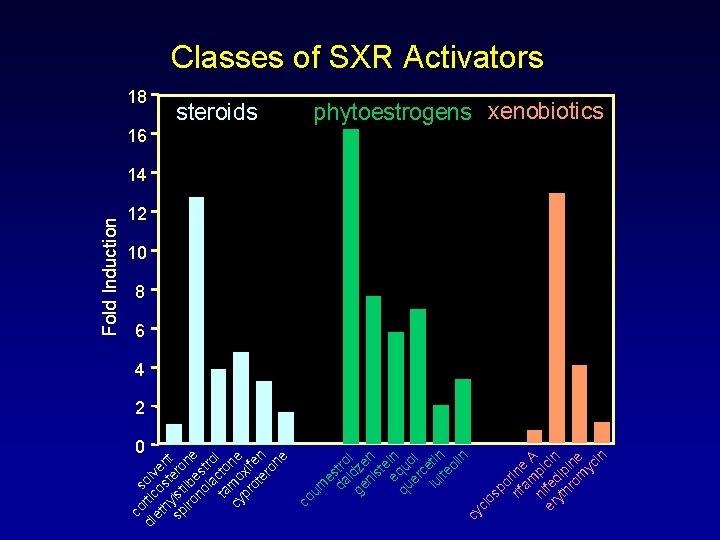
co s di rtic olv et o e h st nt sp ylst ero ilb ne no est la ro ta cto l cy m o ne pr xif ot en er on e 0 steroids cy clo sp o rif rine am A n p er ifed ic in yt ip hr in om e yc in 18 co um e da stro i l ge dze ni n st e eq in qu u er ol c lu etin l te ol in Fold Induction Classes of SXR Activators phytoestrogens xenobiotics 16 14 12 10 8 6 4 2
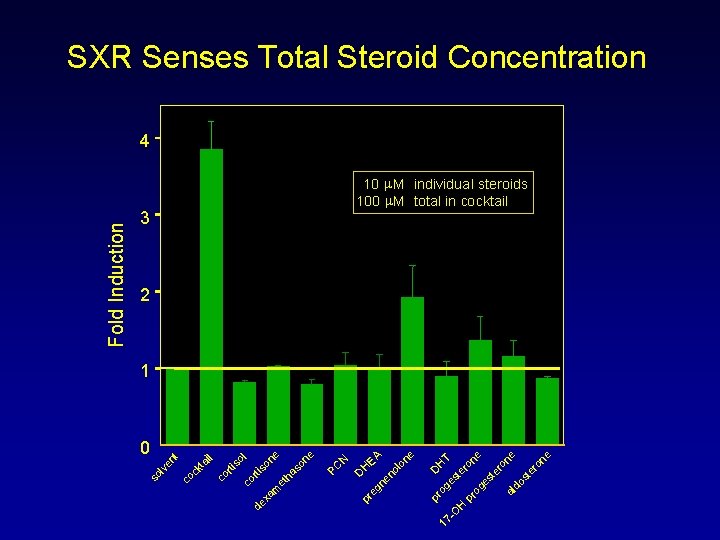
17 - DH og es T O te H ro pr ne og es te ro ne al do st er on e pr D H EA eg ne no lo ne N 3 pr PC il co rt i so l co rt i de so xa ne m et ha so ne ck ta nt 0 co so l ve Fold Induction SXR Senses Total Steroid Concentration 4 10 m. M individual steroids 100 m. M total in cocktail 2 1
Predictions and Requirements of the Model • Sensor should be expressed in tissues that catabolize steroids and xenobiotics – Expressed in liver, small and large intestine • Catabolic enzymes should be targets for the sensor – CYP genes are known targets of SXR, in vivo • Compounds known to induce catabolic enzymes should activate the sensor – Majority of known CYP 3 inducers activate SXR • Partially metabolized (reduced) steroids should activate sensor – Mixture of steroids, each at concentrations below that required to activate SXR, will collectively activate SXR-mediated gene expression
CYP 3 A 4 and Human Steroid Metabolism • Steroid levels are tightly regulated. Increased catabolism will lead to ACTH release and upregulated adrenal synthesis – Observation is elevated ACTH, slightly increased circulating steroids – Decreased circulating steroid metabolites • Increased catabolism will be reflected by urinary metabolites – Large increases in urinary steroids caused by rifampicin therapy have led to misdiagnosis of Cushing’s syndrome – VP 16 SXR transgenic mice have drastically elevated urinary steroid metabolite levels • Induction of CYP 3 A 4 should lead to decreases in administered steroid levels – Steroid crisis in Addison’s patients on rifampicin and oral steroids – Pregnancy in rifampicin-treated patients on oral contraceptives
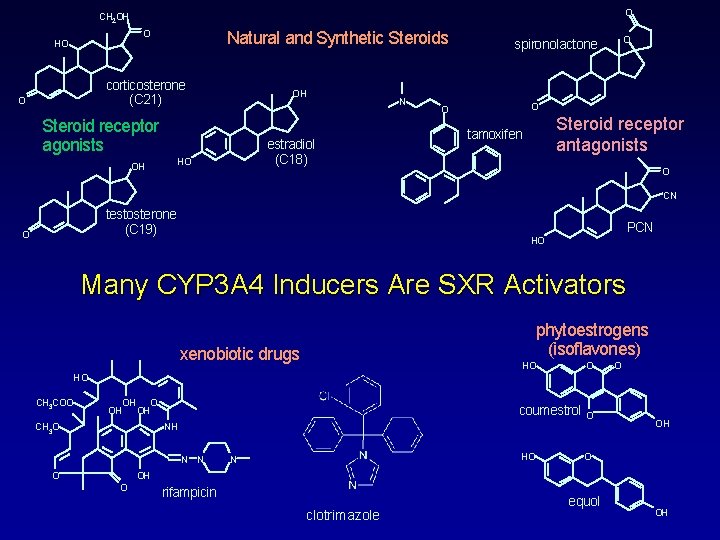
O CH 2 OH O HO Natural and Synthetic Steroids corticosterone (C 21) O OH Steroid receptor agonists estradiol (C 18) HO OH N O spironolactone O O Steroid receptor antagonists tamoxifen O CN testosterone (C 19) O PCN HO Many CYP 3 A 4 Inducers Are SXR Activators phytoestrogens (isoflavones) xenobiotic drugs HO O O HO CH 3 COO OH OH CH 3 O coumestrol O HO O NH N N OH OH O rifampicin clotrimazole equol OH
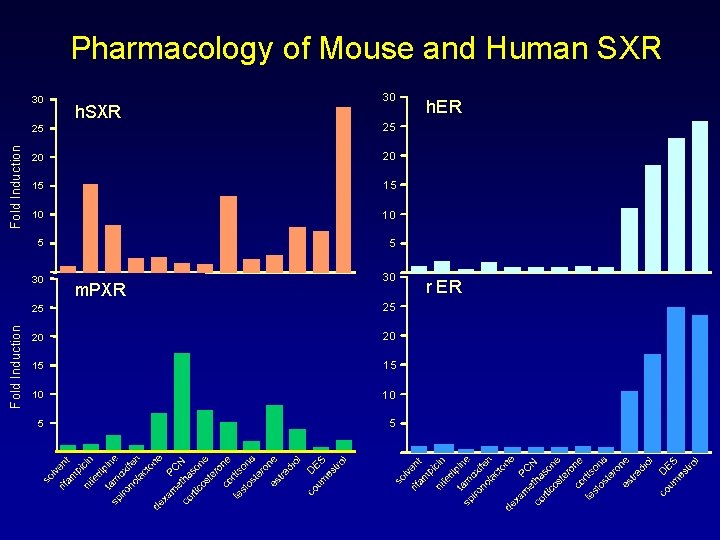
Pharmacology of Mouse and Human SXR Fold Induction 30 h. SXR 30 25 25 20 20 15 15 10 10 5 5 30 30 m. PXR 25 25 20 20 15 15 10 10 5 5 h. ER r ER
Model Systems • Central tenet of model system is parallel biochemistry and endocrinology – Toxicology: effects on animals predict effects on humans – Nuclear receptors behave virtually identically across species • Different pharmacology of SXR and PXR suggests that there are important differences in metabolism • These differences may be highly relevant for toxicology, drug interactions and endocrine disruption • Cross-species extrapolation must account for differences in response of xenobiotic sensors – SXR – CAR
Drug Interactions • CYP 3 A 4 is the primary steroid and xenobiotic metabolizing enzyme • Drugs interactions arise from: – Induction or inhibition of CYP 3 A 4 expression • rifampicin and oral steroids • St John’s Wort and many drugs – Modulation of CYP 3 A 4 enzyme activity • macrolide antibiotics (e. g. erythromycin) and many drugs (e. g. Seldane) • Activated SXR mediates induction of CYP 3 A 4 – SXR activation is a direct molecular test for potential drug interactions • Pharmacological differences between inducibility of rodent and human CYP 3 genes explained by receptor pharmacology – Differences suggest rodents may not be an appropriate model for human drug interactions – Rabbit CYP 3 A induction closely parallels human – Mouse now exists that expresses human SXR instead of mouse gene
SXR and Endocrine Disruption • SXR regulates the P 450 -mediated breakdown of ingested steroids and xenobiotics • Activation of SXR may predict effects of suspected EDC – SXR activators may be detoxified by CYP 3 A action and not a human risk – But activators may also be toxified by CYP 3 A action, increasing the risk. – EDC may have no effect on SXR and therefore more likely to act on other receptors, e. g. ER • SXR is a molecular assay for potential activity of EDCs • Different pharmacology of SXR and PXR suggests that differences in metabolism may exist and be relevant for risk assessment
Approach to Studying EDC Metabolism • Test potential for metabolism by investigating SXR activation by a panel of known and candidate EDCs – Pesticides: DDT, DDE, methoxychlor, endosulfan, dieldrin, alachlor, chlordane, transnonachlor, chlorpyrifos, kepone – Plasticizers: bisphenol A, phthalates – PCBs: e. g. 184, 196 – Alkylphenols: 4 -nonylphenol – xenobiotics: thalidomide, dichlorophenol, triclosan, BHA, BHT • Extend analysis to SXRs from other model organisms of interest – fish - e. g. , zebrafish, medaka, fathead minnow – reptiles - alligator, sea turtle – birds - Japanese quail, zebra finch – amphibians - Xenopus, Rana – mammals - monkey, canine • Investigate actual metabolism in animal models – model organisms including humanized mouse – wild populations
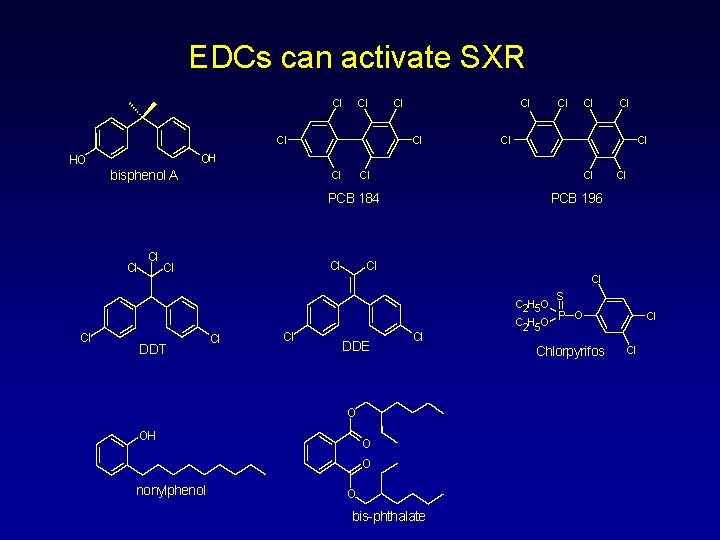
EDCs can activate SXR Cl Cl Cl OH HO bisphenol A Cl Cl Cl PCB 184 Cl Cl Cl DDT PCB 196 Cl Cl DDE Cl O OH O O nonylphenol Cl O bis-phthalate S C 2 H 5 O P O C 2 H 5 O Chlorpyrifos Cl Cl
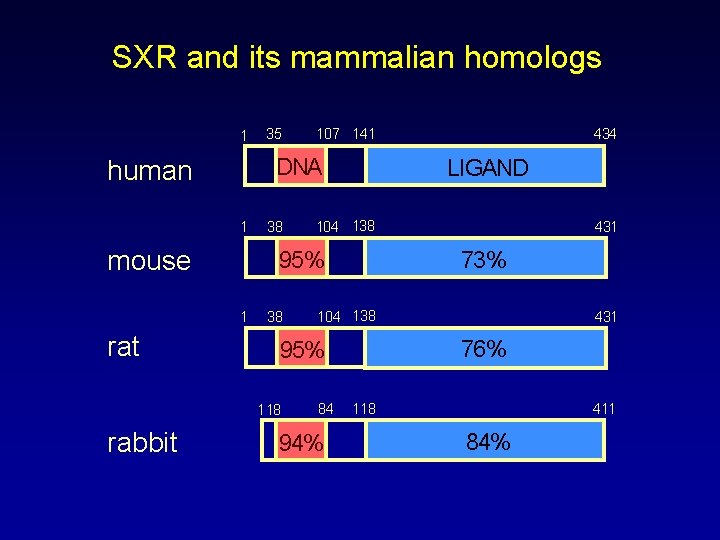
SXR and its mammalian homologs 1 1 mouse 38 38 104 138 431 73% 104 138 84 94% 431 76% 95% 118 434 LIGAND 95% 1 rabbit 107 141 DNA human rat 35 118 411 84%
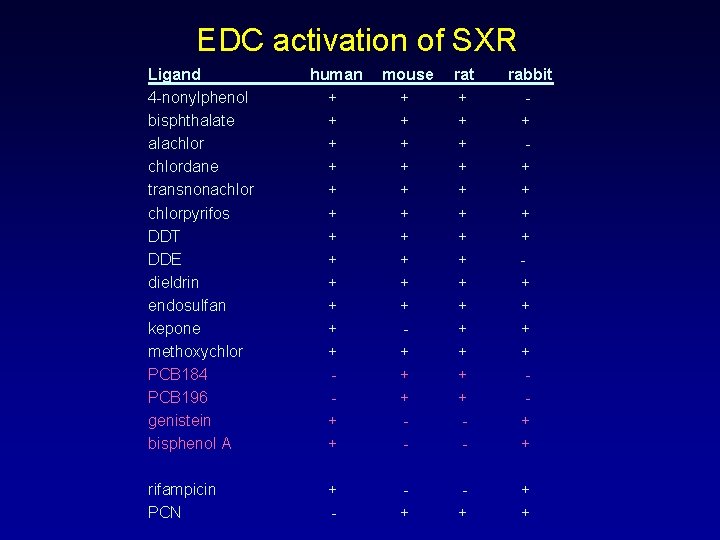
EDC activation of SXR Ligand 4 -nonylphenol bisphthalate alachlordane transnonachlorpyrifos DDT DDE dieldrin endosulfan kepone methoxychlor PCB 184 PCB 196 genistein bisphenol A rifampicin PCN human + + + + mouse + + + + - rat + + + + - rabbit + + + - + +
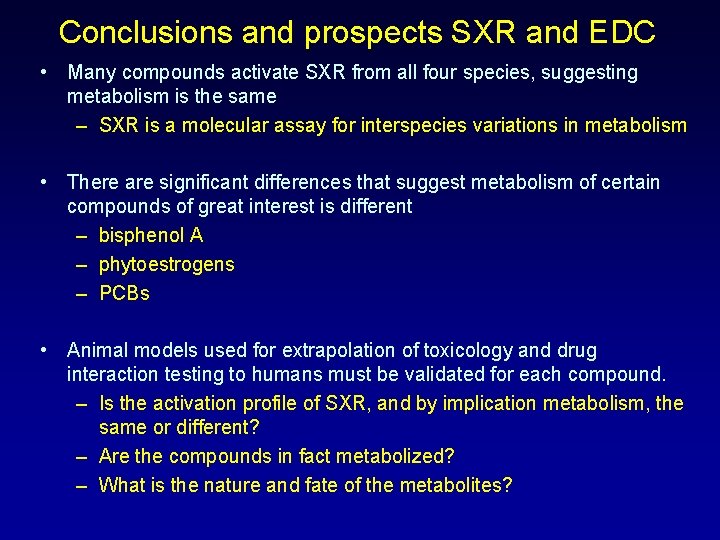
Conclusions and prospects SXR and EDC • Many compounds activate SXR from all four species, suggesting metabolism is the same – SXR is a molecular assay for interspecies variations in metabolism • There are significant differences that suggest metabolism of certain compounds of great interest is different – bisphenol A – phytoestrogens – PCBs • Animal models used for extrapolation of toxicology and drug interaction testing to humans must be validated for each compound. – Is the activation profile of SXR, and by implication metabolism, the same or different? – Are the compounds in fact metabolized? – What is the nature and fate of the metabolites?
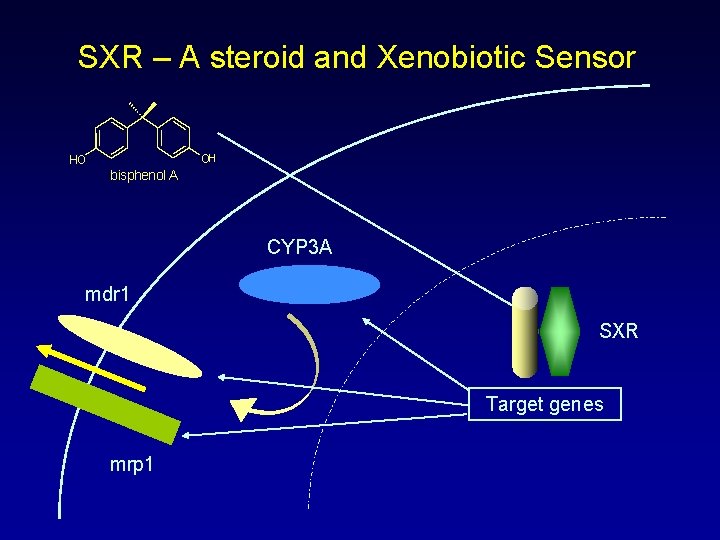
SXR – A steroid and Xenobiotic Sensor OH HO bisphenol A CYP 3 A mdr 1 SXR Target genes mrp 1

SXR - A Steroid and Xenobiotic Sensor • SXR has properties predicted for a steroid and xenobiotic sensor – Expression – Targets – Activators Endogenous and dietary steroids Xenobiotic drugs Environmental toxicants – Expected responses from induction Increased ACTH Increased urinary metabolites Increased circulating steroids but decreased metabolites • SXR is an important molecular test for potential species-specific metabolism of drugs and xenobiotics • Understanding SXR regulation and identifying target genes is an important goal to aid in understanding the xenobiotic response • SXR must be considered when working with model organisms
- Slides: 29