The SQL Select Statement The SQL Select Statement


![SELECT Statement Structure SELECT [field_name(s)] FROM [table_name] WHERE [condition on table row are retrieved] SELECT Statement Structure SELECT [field_name(s)] FROM [table_name] WHERE [condition on table row are retrieved]](https://slidetodoc.com/presentation_image_h/2f8a722e9ae279a41528c90407a3b2e8/image-4.jpg)


![SELECT with Aggregate Function Structure SELECT [field_name(s)] FROM [table_name] WHERE [condition on table row SELECT with Aggregate Function Structure SELECT [field_name(s)] FROM [table_name] WHERE [condition on table row](https://slidetodoc.com/presentation_image_h/2f8a722e9ae279a41528c90407a3b2e8/image-12.jpg)





- Slides: 22

The SQL Select Statement

The SQL Select Statement The select statement is used to select the data from the database and retrieve selected data that match the criteria that you specify. The statement begins with the SELECT keyword. The basic SELECT statement has 3 clauses: SELECT FROM WHERE

The SQL Select Statement The SELECT clause specifies the table field that are retrieved. The FROM clause specifies the tables accessed. The WHERE clause specifies which table rows are retrieved. The WHERE clause is optional; if missing, all table rows are retrieved.
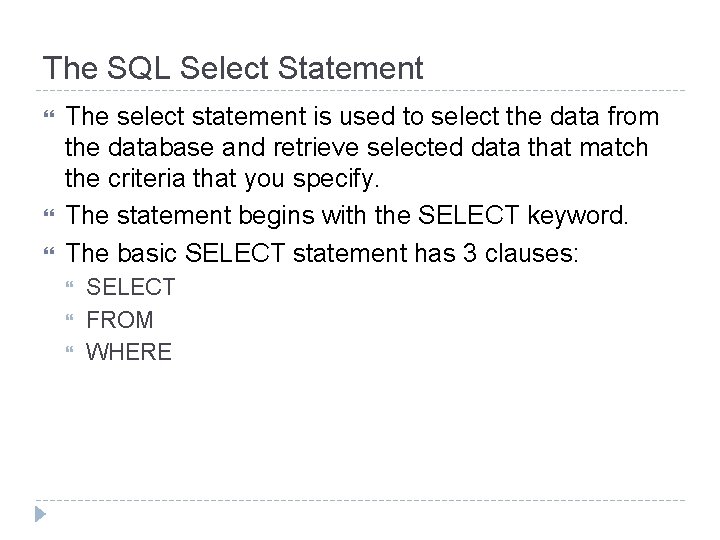
![SELECT Statement Structure SELECT fieldnames FROM tablename WHERE condition on table row are retrieved SELECT Statement Structure SELECT [field_name(s)] FROM [table_name] WHERE [condition on table row are retrieved]](https://slidetodoc.com/presentation_image_h/2f8a722e9ae279a41528c90407a3b2e8/image-4.jpg)
SELECT Statement Structure SELECT [field_name(s)] FROM [table_name] WHERE [condition on table row are retrieved] OR SELECT * FROM [table_name] WHERE [condition on table row are retrieved] Note : SQL is not case sensitive, SELECT is the same as select

Example SELECT * FROM tarbase_nci 60_src_gcrma_u 95 WHERE precursor_micro. RNA = ‘hsa-let-7 a-1’ SELECT clause: select the content of all fields FROM clause: use data from table tarbase_nci 60_scr_gcrma_u 95 WHERE clause: filter those rows where the precursor_micro. RNA field contains hsa-let-7 a-1 Note: we have to use ‘ ‘ (single quote) whenever we state to string; hsa-let-7 a-1 is strin so we have to put it in the single quote like this ‘hsa-let-7 a-1’

Example SELECT * FROM tarbase_nci 60_src_gcrma_u 95 WHERE precursor_micro. RNA LIKE 'hsa-let-7 a%' Select clause: select the content of all fields FROM clause: use the data from tarbase_nci 60_scr_gcrma_u 95 table Where clause: filter those rows where the precursor_micro. RNA field contains the string that start with ‘hsa-let-7 a’, this may return precursor_micro. RNA field including ‘hsa-let-7 a’ or ‘hsa-let-7 a-1’ or ‘hsa-let-7 a-3’ Note: % means whatever presents in this part we ignore it

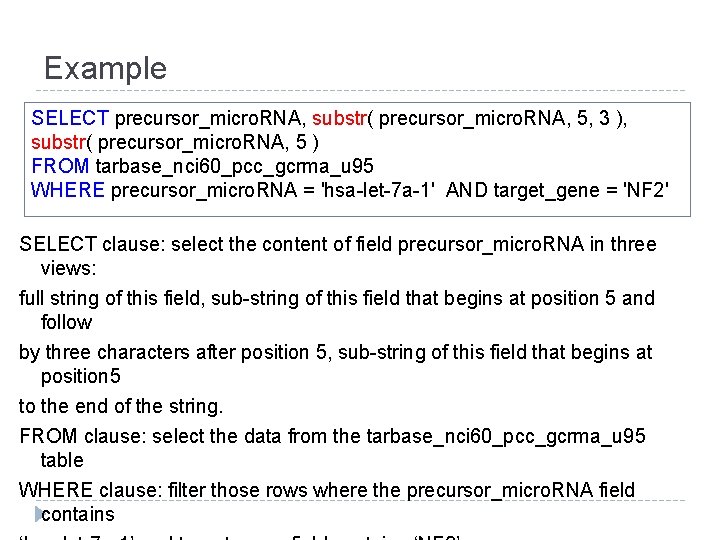
Example SELECT precursor_micro. RNA, substr( precursor_micro. RNA, 5, 3 ), substr( precursor_micro. RNA, 5 ) FROM tarbase_nci 60_pcc_gcrma_u 95 WHERE precursor_micro. RNA = 'hsa-let-7 a-1' AND target_gene = 'NF 2' SELECT clause: select the content of field precursor_micro. RNA in three views: full string of this field, sub-string of this field that begins at position 5 and follow by three characters after position 5, sub-string of this field that begins at position 5 to the end of the string. FROM clause: select the data from the tarbase_nci 60_pcc_gcrma_u 95 table WHERE clause: filter those rows where the precursor_micro. RNA field contains

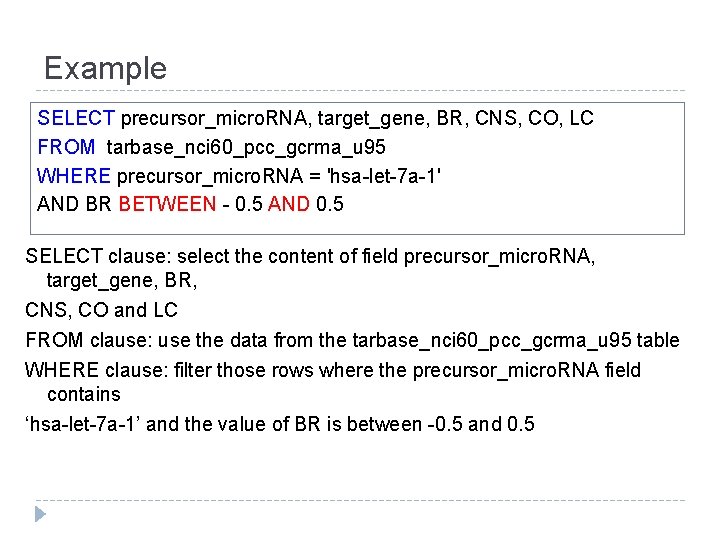
Example SELECT precursor_micro. RNA, target_gene, BR, CNS, CO, LC FROM tarbase_nci 60_pcc_gcrma_u 95 WHERE precursor_micro. RNA = 'hsa-let-7 a-1' AND BR BETWEEN - 0. 5 AND 0. 5 SELECT clause: select the content of field precursor_micro. RNA, target_gene, BR, CNS, CO and LC FROM clause: use the data from the tarbase_nci 60_pcc_gcrma_u 95 table WHERE clause: filter those rows where the precursor_micro. RNA field contains ‘hsa-let-7 a-1’ and the value of BR is between -0. 5 and 0. 5

Example SELECT * FROM tarbase_nci 60_pcc_gcrma_u 95 WHERE precursor_micro. RNA like 'hsa-let-7 a%' and m. RNA_Probe IN ('38007_at', '90076_at', '67793_at’) SELECT clause: select the content of all field FROM clause: select the data from the tarbase_nci 60_pcc_gcrma_u 95 table WHERE clause: filter those rows where the precursor_micro. RNA field contains the string that start with ‘hsa-let-7 a’ and m. RNA_Probe field including ‘ 38007_at’ or ‘ 90076_at’ or ‘ 67793_at’

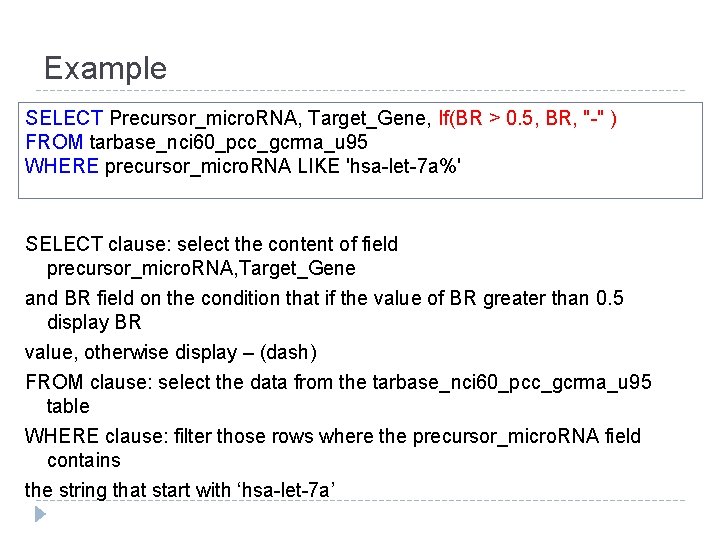
Example SELECT Precursor_micro. RNA, Target_Gene, If(BR > 0. 5, BR, "-" ) FROM tarbase_nci 60_pcc_gcrma_u 95 WHERE precursor_micro. RNA LIKE 'hsa-let-7 a%' SELECT clause: select the content of field precursor_micro. RNA, Target_Gene and BR field on the condition that if the value of BR greater than 0. 5 display BR value, otherwise display – (dash) FROM clause: select the data from the tarbase_nci 60_pcc_gcrma_u 95 table WHERE clause: filter those rows where the precursor_micro. RNA field contains the string that start with ‘hsa-let-7 a’

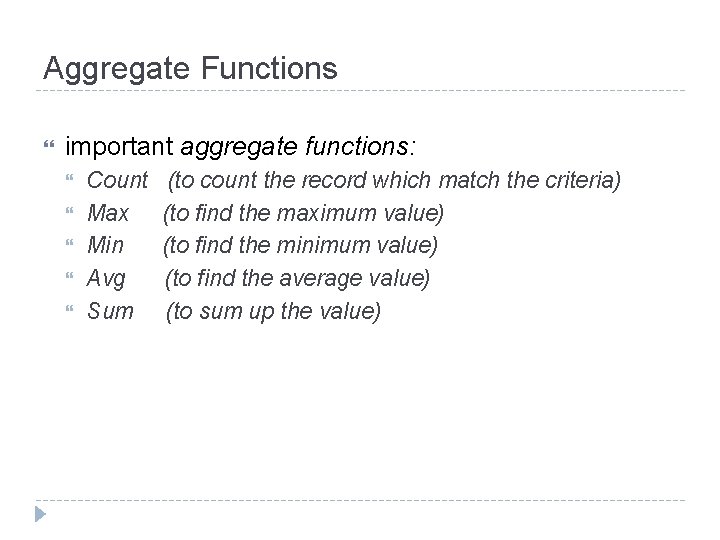
Aggregate Functions important aggregate functions: Count Max Min Avg Sum (to count the record which match the criteria) (to find the maximum value) (to find the minimum value) (to find the average value) (to sum up the value)
![SELECT with Aggregate Function Structure SELECT fieldnames FROM tablename WHERE condition on table row SELECT with Aggregate Function Structure SELECT [field_name(s)] FROM [table_name] WHERE [condition on table row](https://slidetodoc.com/presentation_image_h/2f8a722e9ae279a41528c90407a3b2e8/image-12.jpg)
SELECT with Aggregate Function Structure SELECT [field_name(s)] FROM [table_name] WHERE [condition on table row are retrieved] GROUP BY [field_name(s)] HAVING [condition on grouping field]

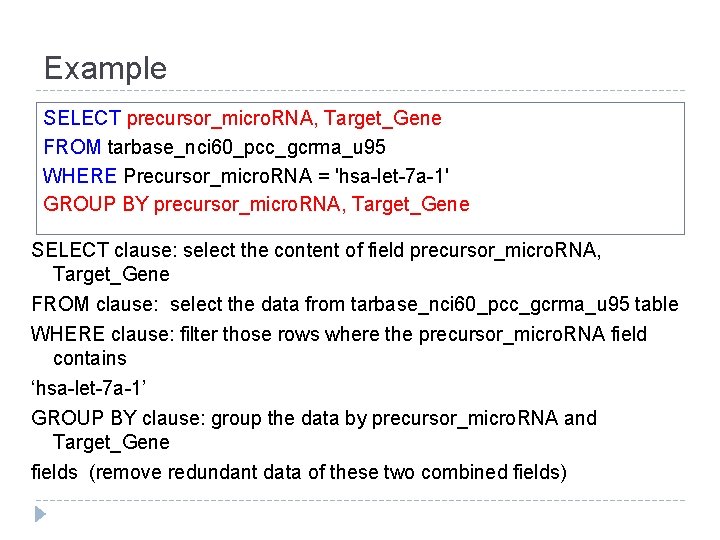
Example SELECT precursor_micro. RNA, Target_Gene FROM tarbase_nci 60_pcc_gcrma_u 95 WHERE Precursor_micro. RNA = 'hsa-let-7 a-1' GROUP BY precursor_micro. RNA, Target_Gene SELECT clause: select the content of field precursor_micro. RNA, Target_Gene FROM clause: select the data from tarbase_nci 60_pcc_gcrma_u 95 table WHERE clause: filter those rows where the precursor_micro. RNA field contains ‘hsa-let-7 a-1’ GROUP BY clause: group the data by precursor_micro. RNA and Target_Gene fields (remove redundant data of these two combined fields)

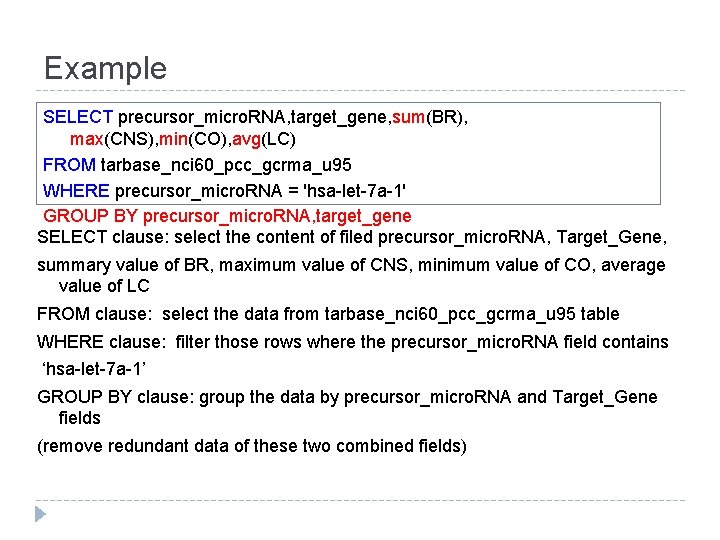
Example SELECT precursor_micro. RNA, target_gene, sum(BR), max(CNS), min(CO), avg(LC) FROM tarbase_nci 60_pcc_gcrma_u 95 WHERE precursor_micro. RNA = 'hsa-let-7 a-1' GROUP BY precursor_micro. RNA, target_gene SELECT clause: select the content of filed precursor_micro. RNA, Target_Gene, summary value of BR, maximum value of CNS, minimum value of CO, average value of LC FROM clause: select the data from tarbase_nci 60_pcc_gcrma_u 95 table WHERE clause: filter those rows where the precursor_micro. RNA field contains ‘hsa-let-7 a-1’ GROUP BY clause: group the data by precursor_micro. RNA and Target_Gene fields (remove redundant data of these two combined fields)

Example SELECT precursor_micro. RNA, target_gene, sum( BR ) , max( CNS ) , min( CO ), avg( LC) FROM tarbase_nci 60_pcc_gcrma_u 95 WHERE precursor_micro. RNA = 'hsa-let-7 a-1' GROUP BY precursor_micro. RNA, target_gene HAVING min( CO ) >0 SELECT clause: select the content of field precursor_micro. RNA , Target_Gene, summation of BR, maximum value of CNS, minimum value of CO, average value of LC FROM clause: select the data from tarbase_nci 60_pcc_gcrma_u 95 table WHERE clause: filter those rows where the precursor_micro. RNA field contains ‘hsa-let-7 a-1’ GROUP BY clause: group the data by precursor_micro. RNA field and Target_Gene field (remove redundant data of these two combined fields) HAVING clause: condition on the CO field, the minimum of CO must be greater than zero

SQL Join SQL join are used to query data from two or more tables, based on a relationship between certain field in these tables. Table in a database are often related to each other with keys. Primary key is a field (or a combination of fields) with a unique value of each row. Each primary key value must be unique within the table. The purpose is to bind data together, across tables, without repeating all the data in every table.

Type of Join Equi JOIN : use only equality comparisons in the jointables. LEFT JOIN : return all rows from the left table, even if there are no matches in the right table. RIGTH JOIN : return all rows from the right table, even if there are no matches in the left table. FULL JOIN : combines the results of both left and right tables. The joined table will contain all records from both tables, and fill in NULLs for missing matches on either side.

Equi Join SELECT a. Precursor_micro. RNA, a. m. RNA_Probe , a. Target_Gene , b. reference, b. pub_date FROM tarbase_nci 60_src_mas 5_u 95 AS a, mirtar AS b WHERE a. precursor_micro. RNA = b. mi. RNA and a. target_gene = b. validated_target and a. precursor_micro. RNA = 'hsa-let-7 g' and a. target_gene = 'HMGA 2' Select the content of field Precursor_micro. RNA, m. RNA_Probe and Target_Gene from table tarbase_nci 60_src_mas 5_u 95, field reference and pub_date from table mirtar Select the data from table tarbase_nci 60_pcc_gcrma_u 95 by using alias name “a” and table mirtar by using alias name “b” Join precursor_micro. RNA field of table “a” to mi. RNA field of table “b” and target_gene field of table “a” with validated_target field of table “b”. And filter those rows where the precursor_micro. RNA field of table “a” contains 'hsa-let-7 g' and target_gene field of table “a” contains 'HMGA 2'

LEFT Join SELECT a. Precursor_micro. RNA , a. m. RNA_Probe , a. Target_Gene, b. reference, b. pub_date FROM tarbase_nci 60_src_mas 5_u 95 AS a LEFT JOIN mirtar AS b ON a. precursor_micro. RNA = b. mi. RNA and a. target_gene = b. validated_target WHERE a. precursor_micro. RNA = 'hsa-let-7 a-1' AND target_gene = 'NF 2' Select the content of field Precursor_micro. RNA, m. RNA_Probe and Target_Gene from table tarbase_nci 60_src_mas 5_u 95, field reference and pub_date from table mirtar Select the data from table tarbase_nci 60_src_gcrma_u 95 by using alias name “a” LEFT JOIN with table mirtar by using alias name “b”. Join precursor_micro. RNA field of table “a” with mi. RNA field of table “b” and target_gene field of table “a” with validated_target field of table “b” Filter those rows where the precursor_micro. RNA field of table “a” contains 'hsa-let-7 a-1‘ and target_gene field of table “a” contains 'NF 2'

Join Table with Query SELECT a. *, b. reference, b. pub_date FROM tarbase_nci 60_src_gcrma_u 95 as a LEFT JOIN (SELECT * FROM mirtar, mi. RNA_precursor_mapping WHERE mirtar. mi. RNA = mi. RNA_precursor_mapping. precursor) as b ON a. precursor_micro. RNA = b. precursor_01 and a. target_gene = b. validated_target WHERE (a. precursor_micro. RNA = 'hsa-let-7 a-3' and a. target_gene = 'NF 2') Select the content of all fields from table tarbase_nci 60_scr_gcrma_u 95 and field reference and pub_date from table mirtar Select the data from table tarbase_nci 60_pcc_gcrma_u 95 by using alias name “a” LEFT JOIN with query which select the content all fields from table mirtar and mi. RNA_precursor_mapping by joining micro. RNA field of table mirtar with precursor field of table mi. RNA_precursor_mapping using alias name “b” Table “a” and the query “b”, join precursor_micro. RNA field of table “a”with precursor_01 field of query “b” and join target_gene field of table “a” with validated_target field of query “b”

Join 3 Tables SELECT a. precursor_micro. RNA, a. target_gene, a. BR, b. reference, b. pub_date, c. MIM_ID FROM tarbase_nci 60_src_gcrma_u 95 AS a, mirtar AS b, tarbase_omim as c WHERE a. precursor_micro. RNA = b. mi. RNA and a. target_gene = b. validated_target and b. validated_target = c. Gene_Symbol and a. precursor_micro. RNA = 'hsa-let. Select 7 g' the content of field precursor_micro. RNA, target_gene, BR from table tarbase_nci 60_src_gcrma_u 95, field reference and pub_date from table mirtar and field MIM_ID from tarbase_omim Select the data from table tarbase_nci 60_src_gcrma_u 95 by using alias name “a”, data from table mirtar by using alias name “b” and data from table tarbase_omim by using alias name “c” WHERE Clause : - join table “a” with table “b” by mapping field precursor_micro. RNA of table “a” with field mi. RNA of table “b” and map field target_gene of table “a” with field validated_target of table “b”. - join table “b” with table “c” by mapping field validated_target of table “b” with field Gene_Symbol of table “c”

Create Table from Query Create Table AAA as SELECT a. precursor_micro. RNA, a. target_gene, a. BR, b. reference, b. pub_date, c. MIM_ID FROM tarbase_nci 60_src_gcrma_u 95 AS a, mirtar AS b, tarbase_omim as c WHERE a. precursor_micro. RNA = b. mi. RNA and a. target_gene = b. validated_target and b. validated_target = c. Gene_Symbol and a. precursor_micro. RNA = 'hsa-let-7 g' Create clause : create static table name AAA from following query in the database