The role of enamel matrix proteins in enamel






- Slides: 37

The role of enamel matrix proteins in enamel morphogenesis and mineralisation Maria Augusta Landin 2011

• Enamel formation • Enamel matrix proteins • Protein structure • Splicing products • Degradation • Function/role during enamel formation ∙ morphogenesis ∙ mineralization • Summary -In other mineralized tissues -Clinical importance and use 2011

Enamel matrix formation • Early secretory/presecretory stage -Preameloblast elongation (cylinder shape) -Polarization • Secretory stage - Tomes process Secretion of enamel proteins Cleavage/proteolysis Transport of processed enamel protein out of the ameloblasts Hydroxyapatite crystals formation • Maturation stage -The enamel proteins are digested by proteinases and transported from the enamel matrix into the ameloblasts -Hydroxyapatite cristalls are transported out of the ameloblasts into the matrix - Enamel crystals grow in length and width - Ameloblasts die by apoptosis 2011

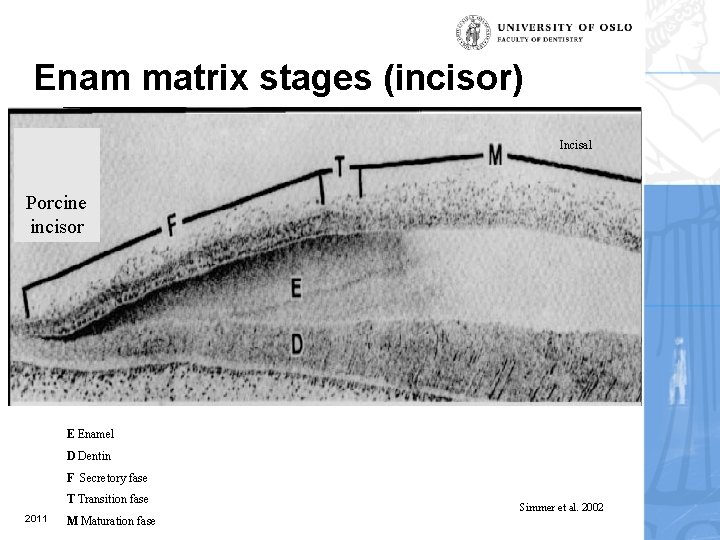
Enam matrix stages (incisor) Incisal Porcine incisor E Enamel D Dentin F Secretory fase T Transition fase 2011 M Maturation fase Simmer et al. 2002

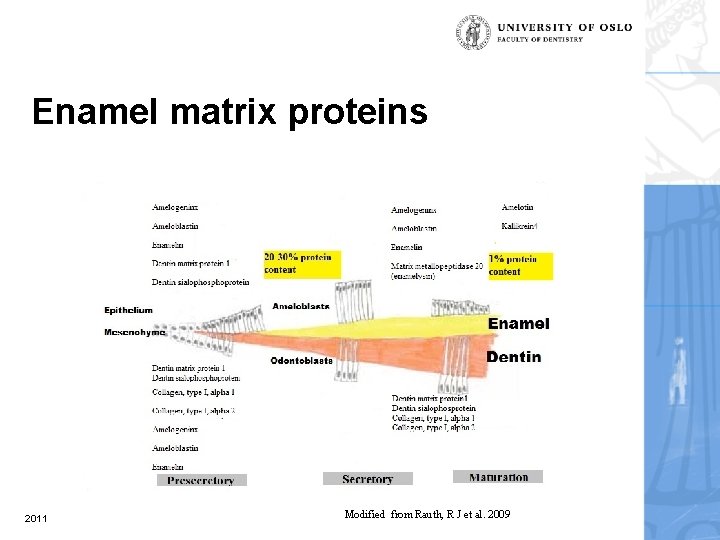
Enamel matrix proteins 2011 Modified from Rauth, R J et al. 2009

Enamel matrix & matrix proteins • The organic matrix of the developing enamel is highly heterogeneous, e. g. amelogenin, enamelin, ameloblastin, tuftelin, dentine sialophosphoprotein, amelotin, biglycan, enzymes (encoded by Klk 4 and Mmp 20) and serum proteins. 2011

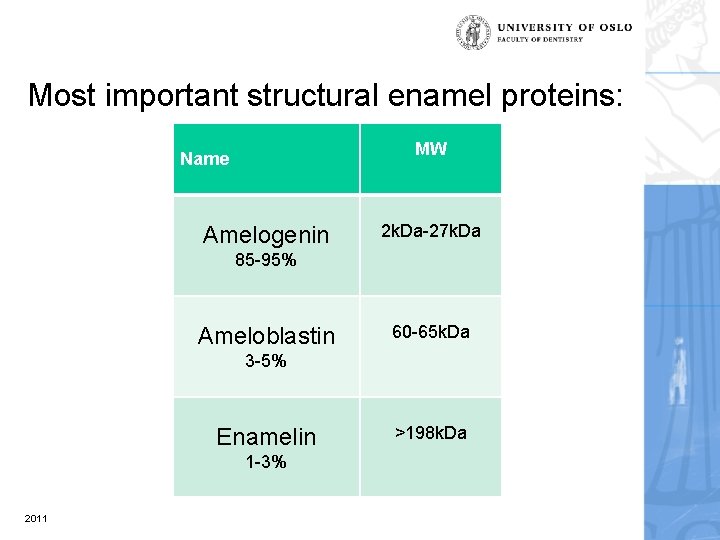
Most important structural enamel proteins: MW Name Amelogenin 2 k. Da-27 k. Da 85 -95% Ameloblastin 60 -65 k. Da 3 -5% Enamelin 1 -3% 2011 >198 k. Da

• Each of these proteins undergo post-secretory sequential degradation which contributes towards further matrix heterogeneity. • These proteins play an important role in the modulation of mineral deposition and growth during tooth morphogenesis. 2011

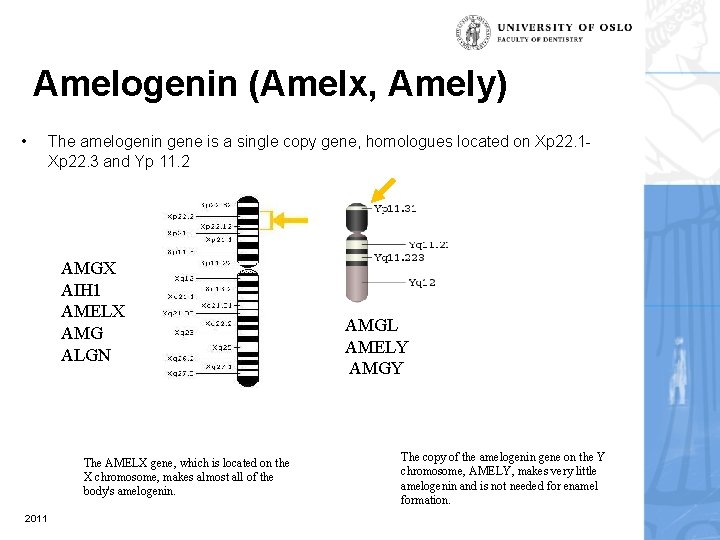
Amelogenin (Amelx, Amely) • The amelogenin gene is a single copy gene, homologues located on Xp 22. 1 Xp 22. 3 and Yp 11. 2 AMGX AIH 1 AMELX AMG ALGN The AMELX gene, which is located on the X chromosome, makes almost all of the body's amelogenin. 2011 AMGL AMELY AMGY The copy of the amelogenin gene on the Y chromosome, AMELY, makes very little amelogenin and is not needed for enamel formation.

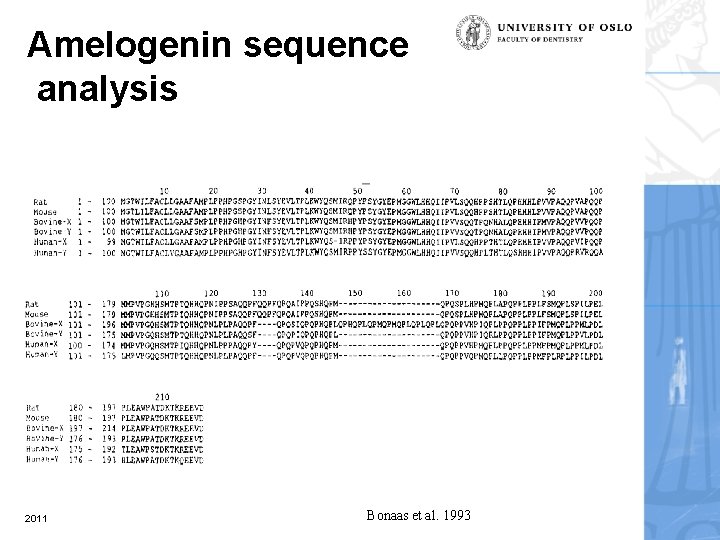
Amelogenin sequence analysis 2011 Bonaas et al. 1993

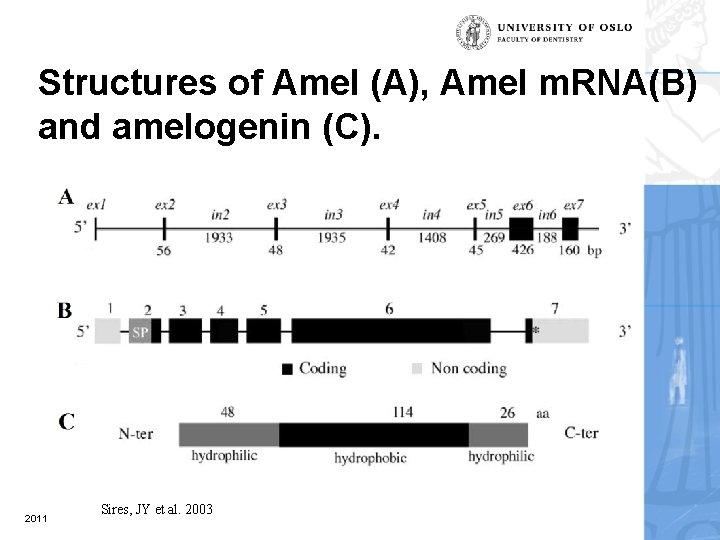
Structures of Amel (A), Amel m. RNA(B) and amelogenin (C). 2011 Sires, JY et al. 2003

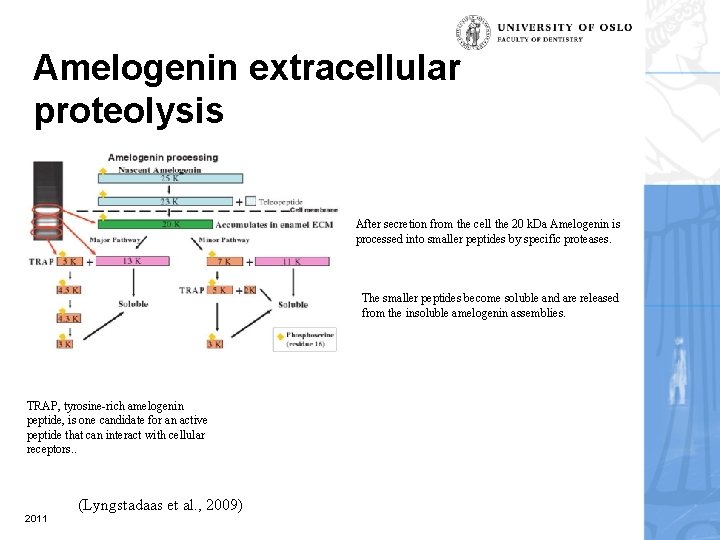
Amelogenin extracellular proteolysis After secretion from the cell the 20 k. Da Amelogenin is processed into smaller peptides by specific proteases. The smaller peptides become soluble and are released from the insoluble amelogenin assemblies. TRAP, tyrosine-rich amelogenin peptide, is one candidate for an active peptide that can interact with cellular receptors. . 2011 (Lyngstadaas et al. , 2009)

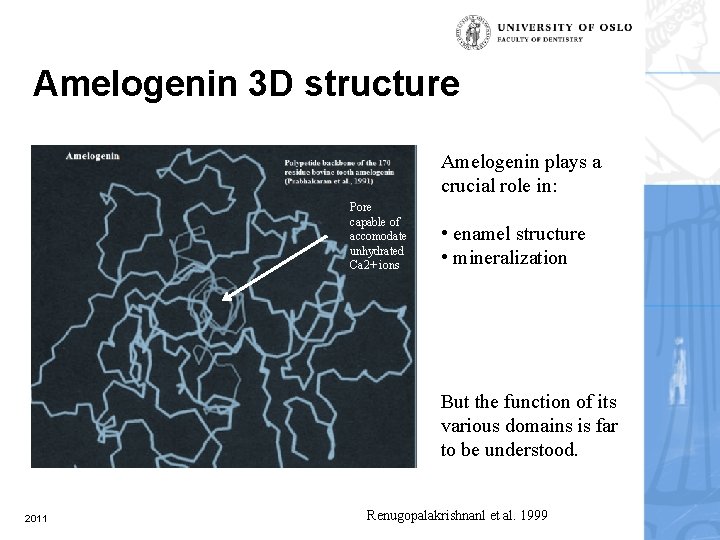
Amelogenin 3 D structure Amelogenin plays a crucial role in: Pore capable of accomodate unhydrated Ca 2+ ions • enamel structure • mineralization But the function of its various domains is far to be understood. 2011 Renugopalakrishnanl et al. 1999

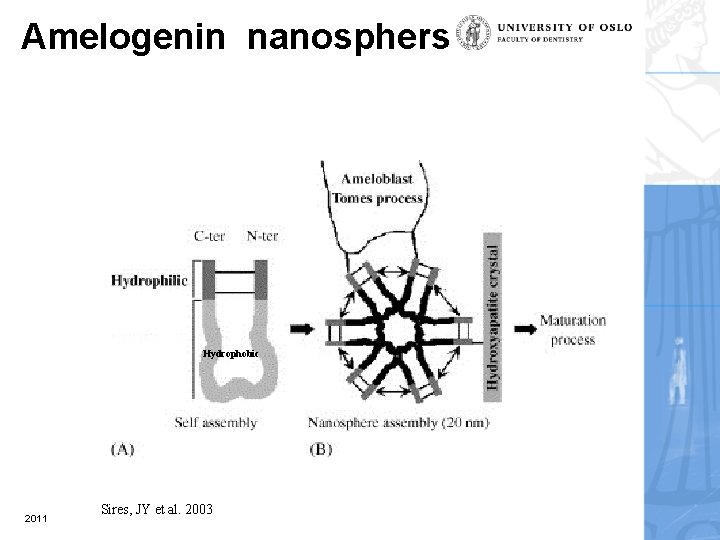
Amelogenin nanosphers Hydrophobic 2011 Sires, JY et al. 2003

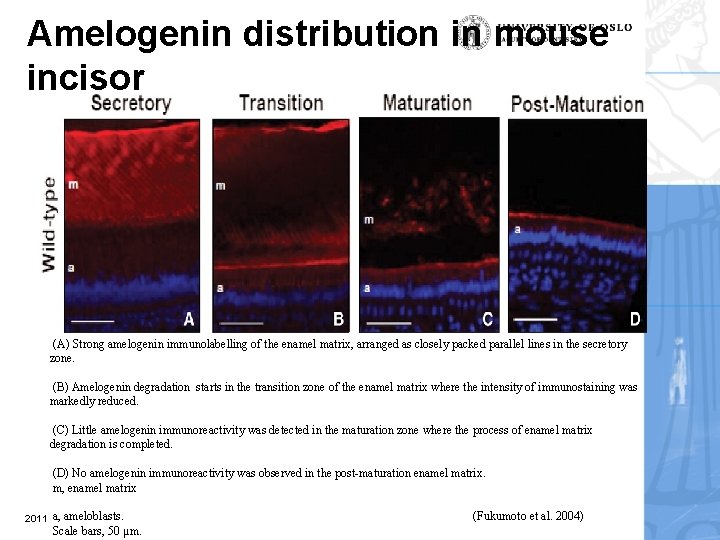
Amelogenin distribution in mouse incisor (A) Strong amelogenin immunolabelling of the enamel matrix, arranged as closely packed parallel lines in the secretory zone. (B) Amelogenin degradation starts in the transition zone of the enamel matrix where the intensity of immunostaining was markedly reduced. (C) Little amelogenin immunoreactivity was detected in the maturation zone where the process of enamel matrix degradation is completed. (D) No amelogenin immunoreactivity was observed in the post-maturation enamel matrix. m, enamel matrix 2011 a, ameloblasts. Scale bars, 50 µm. (Fukumoto et al. 2004)

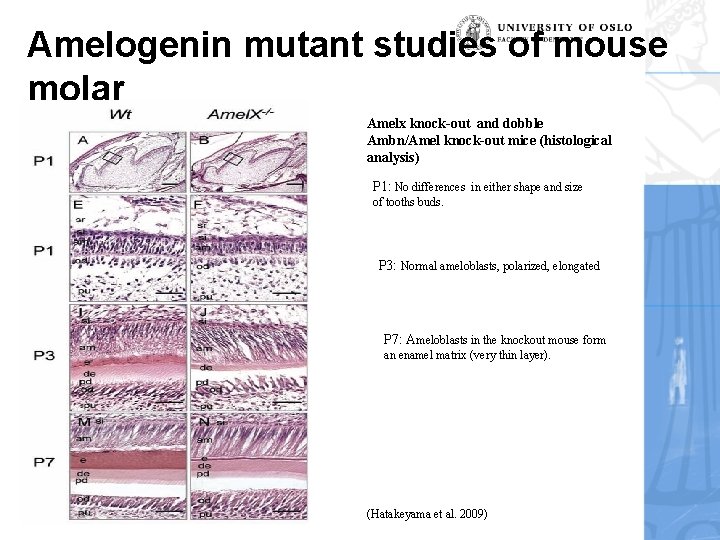
Amelogenin mutant studies of mouse molar Amelx knock-out and dobble Ambn/Amel knock-out mice (histological analysis) P 1: No differences in either shape and size of tooths buds. P 3: Normal ameloblasts, polarized, elongated P 7: Ameloblasts in the knockout mouse form an enamel matrix (very thin layer). 2011 (Hatakeyama et al. 2009)

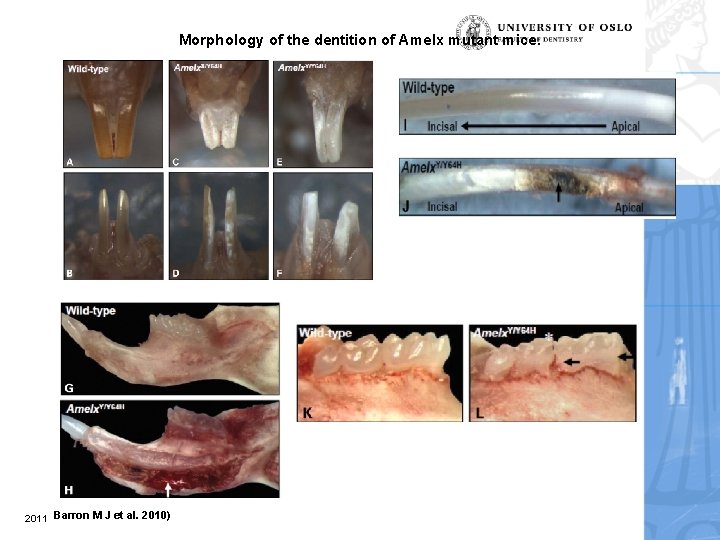
Morphology of the dentition of Amelx mutant mice. 2011 Barron M J et al. 2010)

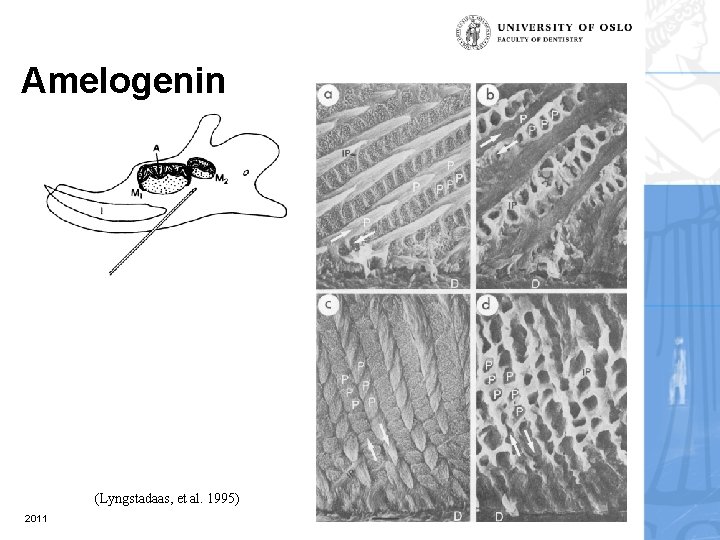
Amelogenin (Lyngstadaas, et al. 1995) 2011

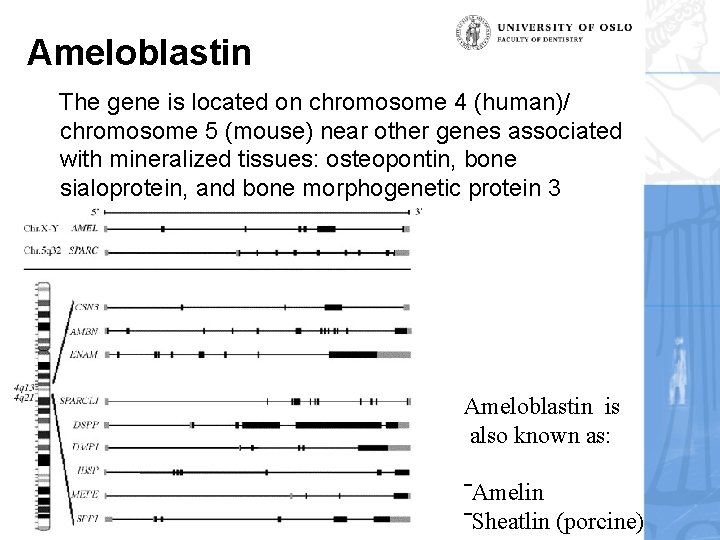
Ameloblastin The gene is located on chromosome 4 (human)/ chromosome 5 (mouse) near other genes associated with mineralized tissues: osteopontin, bone sialoprotein, and bone morphogenetic protein 3 Ameloblastin is also known as: 2011 ‾Amelin ‾Sheatlin (porcine)

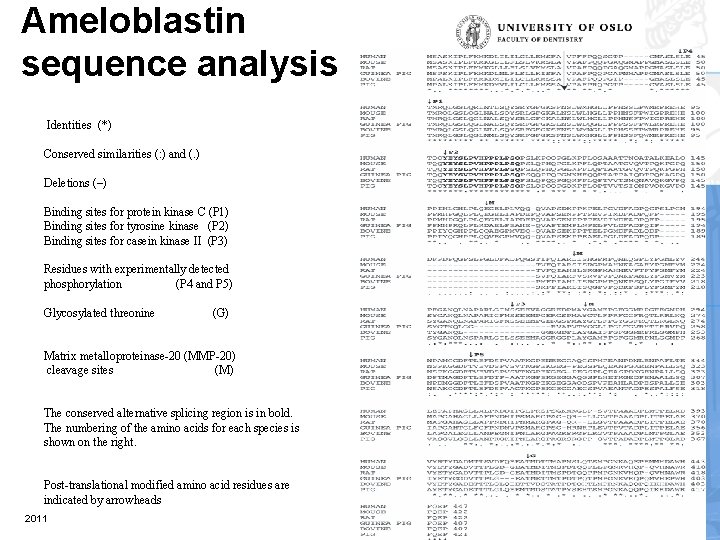
Ameloblastin sequence analysis Identities (*) Conserved similarities (: ) and (. ) Deletions (–) Binding sites for protein kinase C (P 1) Binding sites for tyrosine kinase (P 2) Binding sites for casein kinase II (P 3) Residues with experimentally detected phosphorylation (P 4 and P 5) Glycosylated threonine (G) Matrix metalloproteinase-20 (MMP-20) cleavage sites (M) The conserved alternative splicing region is in bold. The numbering of the amino acids for each species is shown on the right. Post-translational modified amino acid residues are indicated by arrowheads 2011

Ameloblastin Structural three-dimensional (3 D) model of human ameloblastin 2011 Vimetal, J et al. 2008

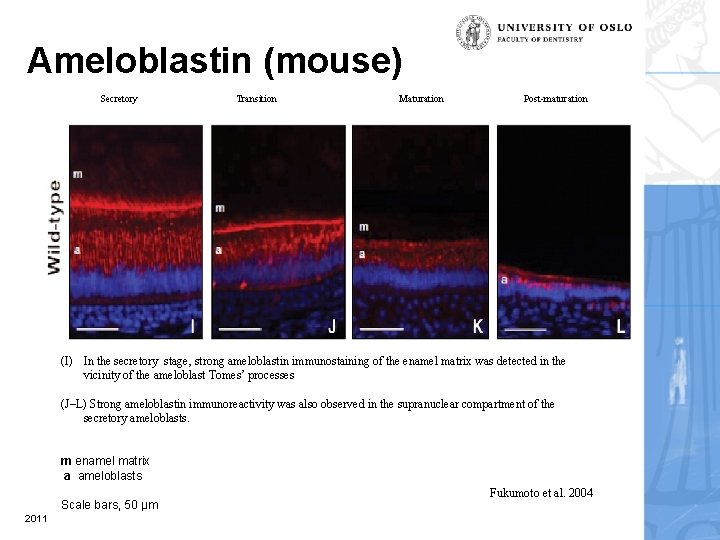
Ameloblastin (mouse) Secretory Transition Maturation Post-maturation (I) In the secretory stage, strong ameloblastin immunostaining of the enamel matrix was detected in the vicinity of the ameloblast Tomes’ processes (J–L) Strong ameloblastin immunoreactivity was also observed in the supranuclear compartment of the secretory ameloblasts. m enamel matrix a ameloblasts Scale bars, 50 µm 2011 Fukumoto et al. 2004

Ameloblastin (mutant studies) Ambn knock-out and dobble Ambn/Amel knock-out mice (histological analysis) P 1: No differences in either shape and size of tooths buds. P 3: Ameloblasts detach from the matrix layer and loose cell polarity. P 7: Ameloblasts completly loose cell polarity and accumulate to form a multilayred structure. The Ambn mutant contained calcified nodules (arrow) Fukumoto S et al. 2004 2011 Hatakeyama et al. 2009

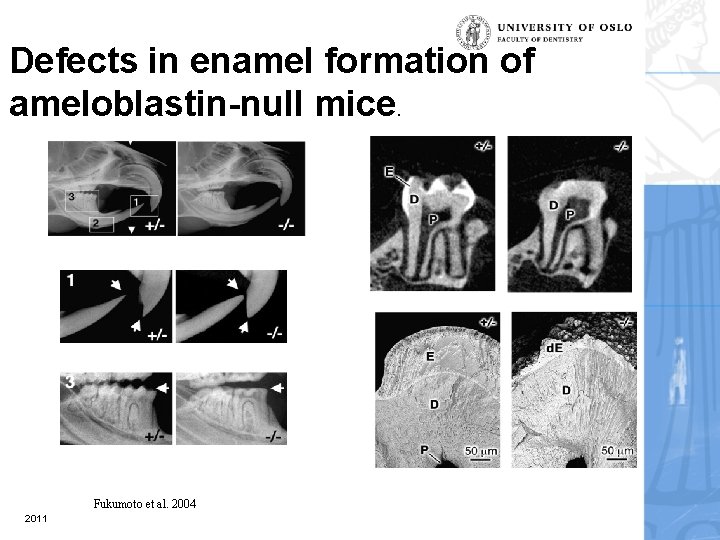
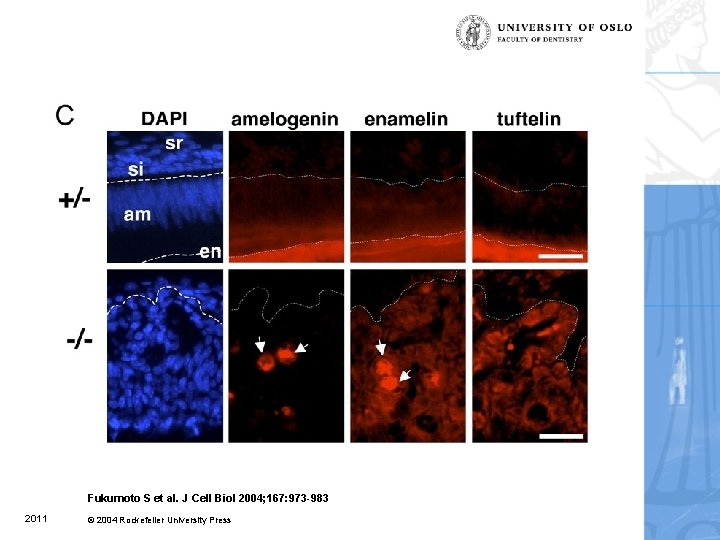
Defects in enamel formation of ameloblastin-null mice. Fukumoto et al. 2004 2011

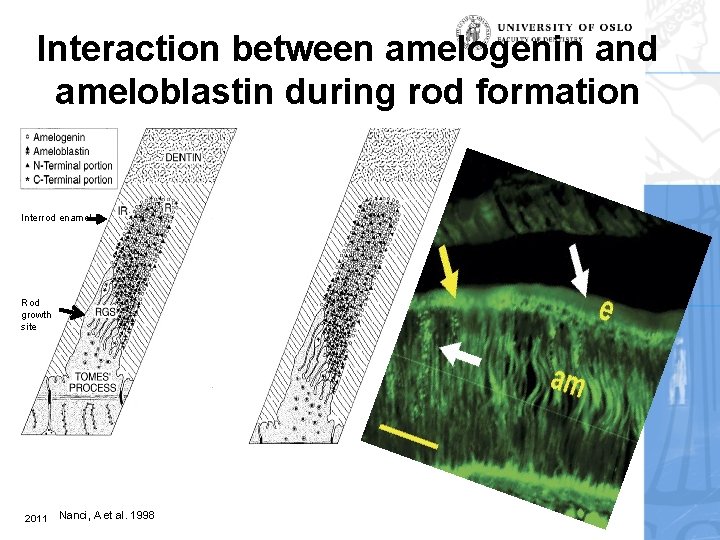
Interaction between amelogenin and ameloblastin during rod formation Interrod enamel Rod growth site 2011 Nanci, A et al. 1998

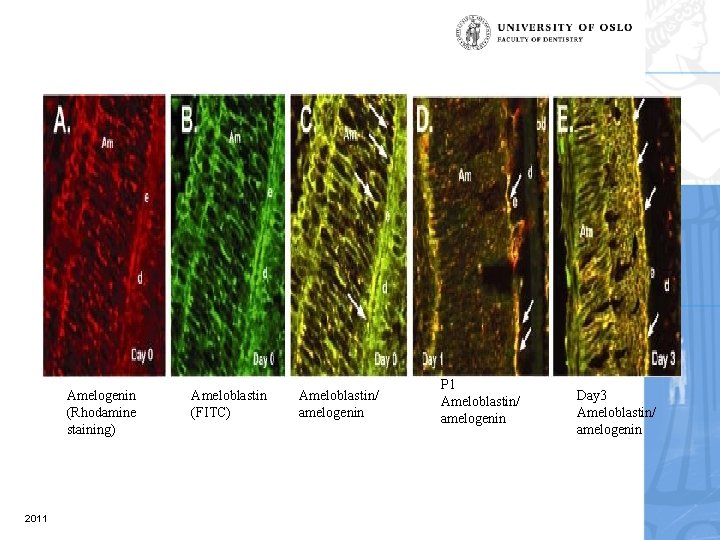
Amelogenin (Rhodamine staining) 2011 Ameloblastin (FITC) Ameloblastin/ amelogenin P 1 Ameloblastin/ amelogenin Day 3 Ameloblastin/ amelogenin

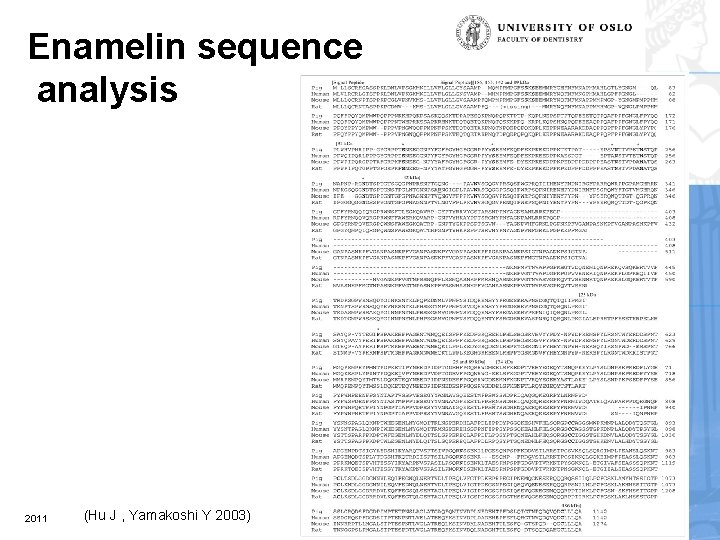
Enamelin sequence analysis 2011 (Hu J , Yamakoshi Y 2003)

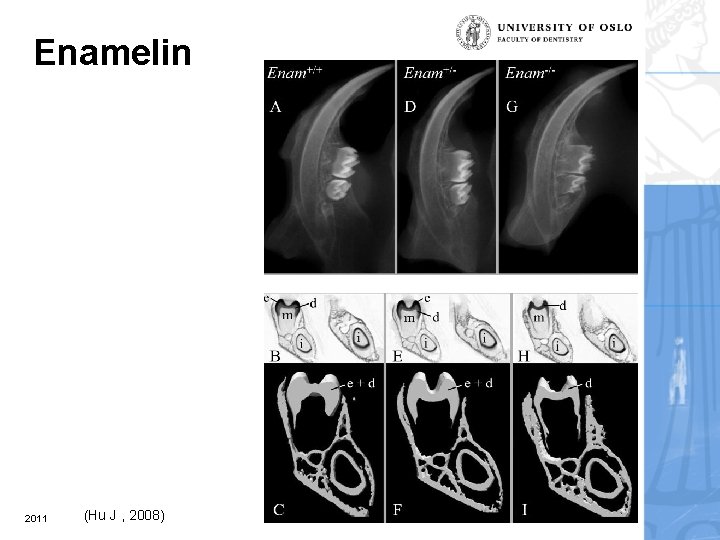
Enamelin 2011 (Hu J , 2008)

Enamelin 2011 (Hu J , 2008)

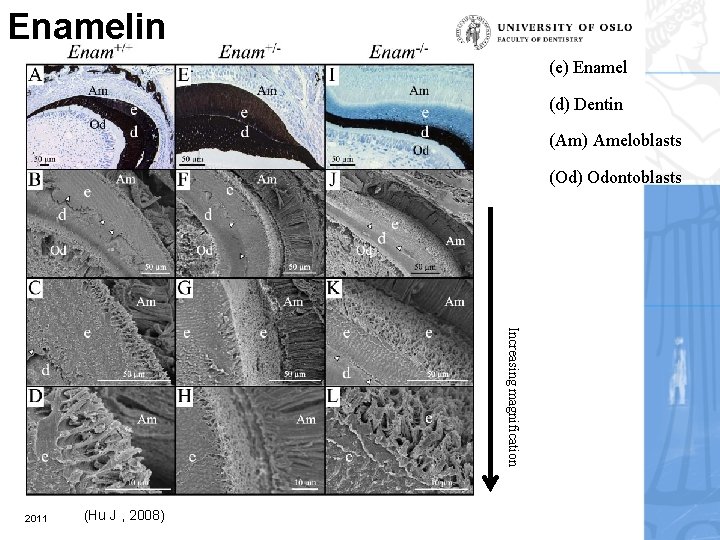
Enamelin (e) Enamel (d) Dentin (Am) Ameloblasts (Od) Odontoblasts Increasing magnification 2011 (Hu J , 2008)

Interaction between amelogenin and enamelin http: //www. sciencedirect. com/science? _ob=Article. URL&_udi=B 6 WM 5 -4 VDH 8 WM 5&_user=10&_cover. Date=04%2 F 30%2 F 2009&_rdoc=1&_fmt=high&_orig=gateway&_origin=gateway&_sort=d&_doc anchor=&view=c&_search. Str. Id=1687732364&_rerun. Origin=google&_acct=C 000050221&_version=1&_url. Version=0 &_userid=10&md 5=f 9 ac 3767 fa 72816 f 73 a 92 ab 8 bf 8 e 73 c 8&searchtype=a In vitro study on the interaction between the 32 k. Da enamelin and amelogenin. Daming Fana, Chang Dua, Zhi Suna, Rajamani Lakshminarayanana and Janet Moradian-Oldak J structural biology 2009; 166: 88 cooperation between enamelin and amelogenin in macromolecular self-assembly and in controlling enamel mineral formation 2011

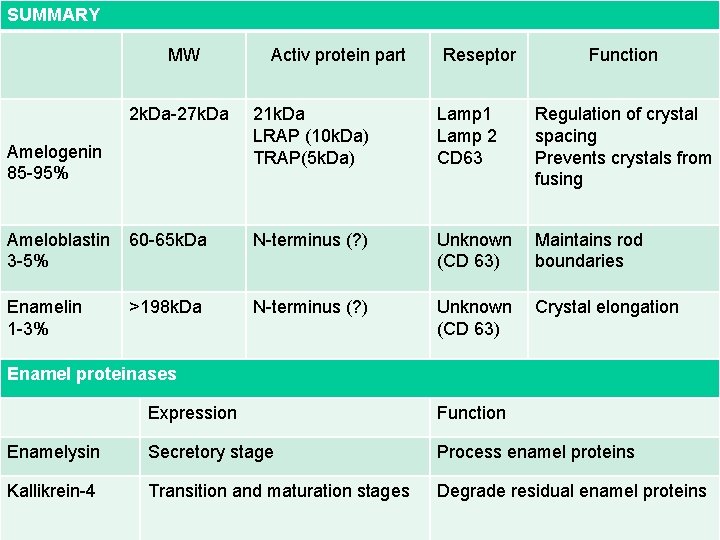
SUMMARY MW Activ protein part Reseptor Function 2 k. Da-27 k. Da 21 k. Da LRAP (10 k. Da) TRAP(5 k. Da) Lamp 1 Lamp 2 CD 63 Regulation of crystal spacing Prevents crystals from fusing Ameloblastin 3 -5% 60 -65 k. Da N-terminus (? ) Unknown (CD 63) Maintains rod boundaries Enamelin 1 -3% >198 k. Da N-terminus (? ) Unknown (CD 63) Crystal elongation Amelogenin 85 -95% Enamel proteinases Expression Function Enamelysin Secretory stage Process enamel proteins Kallikrein-4 Transition and maturation stages Degrade residual enamel proteins 2011

Thank you for your attention! 2011

Other functions of ameloblastin • Signal molecule in epithelial-mesenchymal interactions • Promotes the differentiation and growth of mesenchymal cells • Ambn is also expressed during the development of mesenchymal dental tissue ( dentin ) and pulp. • During trauma-induced reparative dentin formation • During early stages of craniofacial bone formation • Induces hard-tissue regeneration • Wound healing 2011

Ameloblastin Gene Ontology (GO) • The gene products of Ambn are associated mainly with: − − − Structural constituent of tooth enamel Odontogenesis of dentine Growth factor activity Cell adhesion Cell proliferation Bone mineralization Sett inn linken…. . 2011

Fukumoto S et al. J Cell Biol 2004; 167: 973 -983 2011 © 2004 Rockefeller University Press

Summary • Ameloblastin ∙ Essential for maintaining the differentiation state of the ameloblasts ∙ Importante cell adhesion molecule ∙ Plays a central rolle in mineralisation. Ambn knockout mouse show hypoplasy (thin enamel) ∙ 2011 Required for normal tooth morphology