The polymerase chain reaction PCR Experiment Goals Understand






- Slides: 26

The polymerase chain reaction (PCR)

Experiment Goals • Understand how PCR technique works. • Perform PCR experiment. • Analyze PCR products.

What is PCR? Definition • Is a powerful and sensitive technique to amplify a piece of DNA very rapidly outside a living cell (DNA amplification in vitro).

PCR Applications • PCR is now a common and often indispensable (main) technique used in medical and biological research labs for a variety of applications. Structural analysis Disease detection Cloning Mutation analysis Mapping Sequencing Forensic medicine Scientific research Pre-natal diagnosis

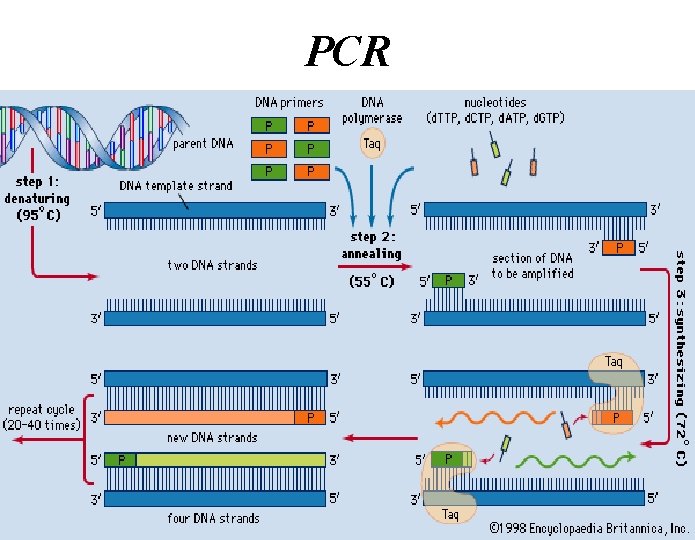
How does PCR work? • There are three major steps in a PCR which are repeated for 30 or 40 cycles. • This is done on an automated cycler, which can heat and cool the tubes with the reaction mixture in a very short time.

Denaturation • The initial step denatures the target DNA (Hbonds are broken between strands of DNA with heat) by heating it to 94°C or higher. • In the denaturation process, the two intertwined (coiled) strands of DNA separate from one another, producing the necessary single-stranded DNA template for replication by thermo stable DNA polymerase.

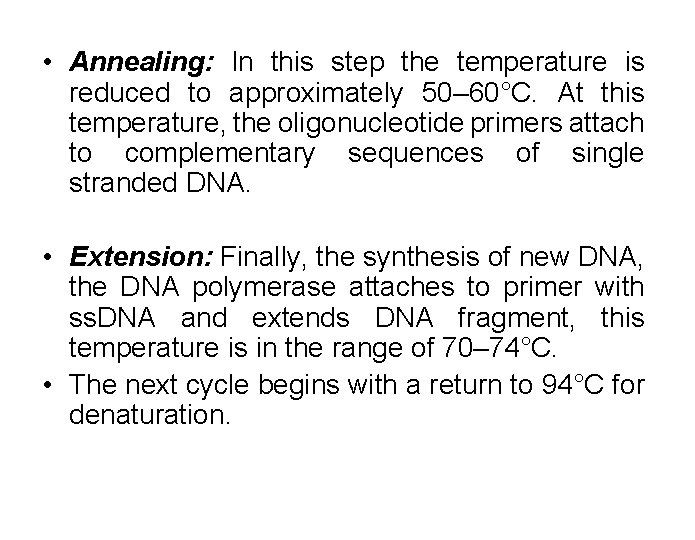
• Annealing: In this step the temperature is reduced to approximately 50– 60°C. At this temperature, the oligonucleotide primers attach to complementary sequences of single stranded DNA. • Extension: Finally, the synthesis of new DNA, the DNA polymerase attaches to primer with ss. DNA and extends DNA fragment, this temperature is in the range of 70– 74°C. • The next cycle begins with a return to 94°C for denaturation.

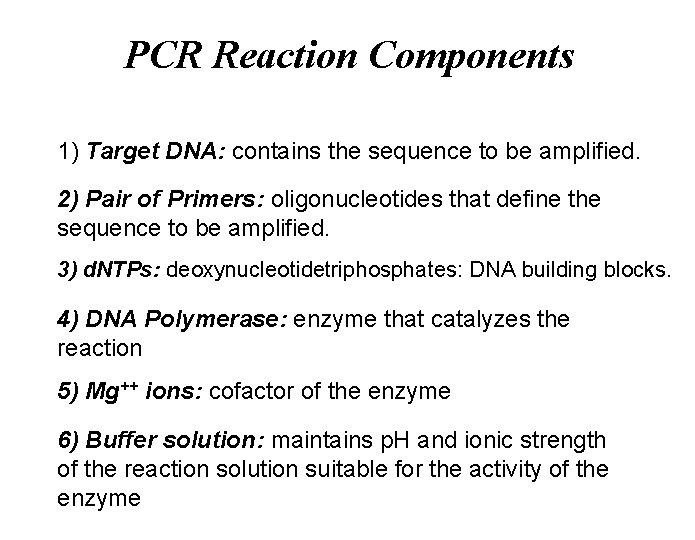
PCR Reaction Components 1) Target DNA: contains the sequence to be amplified. 2) Pair of Primers: oligonucleotides that define the sequence to be amplified. 3) d. NTPs: deoxynucleotidetriphosphates: DNA building blocks. 4) DNA Polymerase: enzyme that catalyzes the reaction 5) Mg++ ions: cofactor of the enzyme 6) Buffer solution: maintains p. H and ionic strength of the reaction solution suitable for the activity of the enzyme

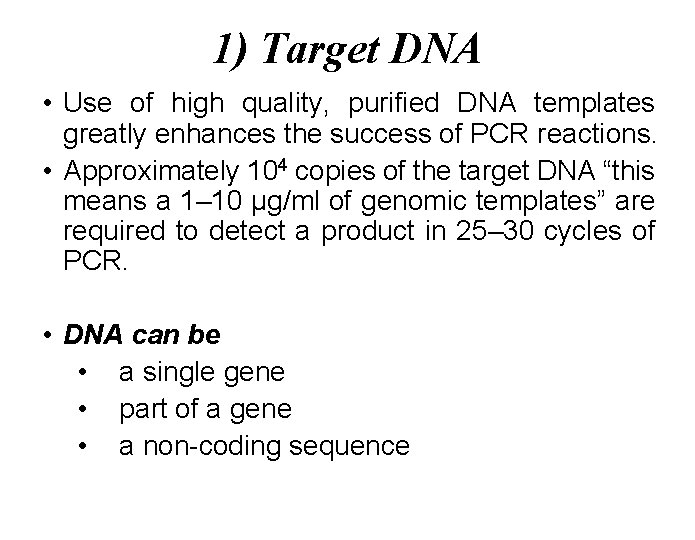
1) Target DNA • Use of high quality, purified DNA templates greatly enhances the success of PCR reactions. • Approximately 104 copies of the target DNA “this means a 1– 10 µg/ml of genomic templates” are required to detect a product in 25– 30 cycles of PCR. • DNA can be • a single gene • part of a gene • a non-coding sequence

DNA Quality • DNA should be intact and free of contaminants that inhibit amplification. – Contaminants can be purified from the original DNA source. • Heme from blood, and melanin from hair – Contaminants can be introduced during the purification process. • Phenol, ethanol, detergents, and salts.

How Big A Target is? • Amplification products are typically in the size range 100 -1500 bp. • Longer targets are amplifiable >25 kb. • Requires modified reaction buffer, cocktails of polymerases, and longer extension times.

2) Pair of Primers • Primers: the short DNA molecules sequence to be amplified. • Oligonucleotide primers • Generally 20– 30 nucleotides in length. • Primers bind (anneal) to the DNA template and act as starting points for the DNA polymerase, – DNA polymerases cannot initiate DNA synthesis without a primer.

3) d. NTPs (deoxynucleotidetriphosphates) – The building blocks for synthesized DNA strands. – d. ATP, d. GTP, d. CTP or d. TTP the newly

4) DNA Polymerase • DNA Polymerase is the enzyme responsible for copying the sequence starting at the primer from the single DNA strand • Commonly use Taq, an enzyme from the hyperthermophilic organisms Thermus aquaticus, isolated first at a thermal spring • This enzyme is heat-tolerant

PCR

Running PCR • The PCR is commonly carried out in a reaction volume of 15 -100 μl in small reaction tubes (0. 2 -0. 5 ml volumes) in a thermal cycler. • The thermal cycler allows heating and cooling of the reaction tubes to control the temperature required at each reaction step. • Most thermal cyclers have heated lids to prevent condensation at the top of the reaction tube. • Older thermocyclers lacking a heated lid require a layer of oil on top of the reaction mixture or a ball of wax inside the tube.

Initialization step • Prior to the first cycle, there is an initialization step – the PCR reaction is often heated to a temperature of 94 -96°C, and this temperature is then held for 1 -9 minutes – This first hold is employed to ensure that most of the DNA template and primers are denatured, – Also, some PCR polymerases require this step for activation

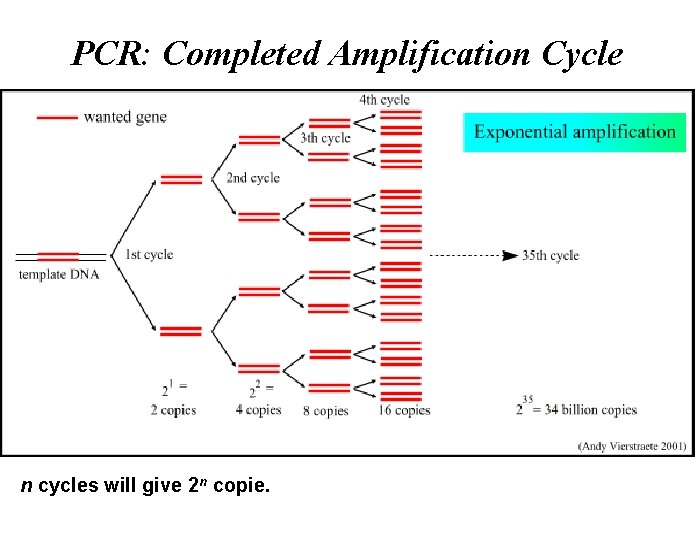
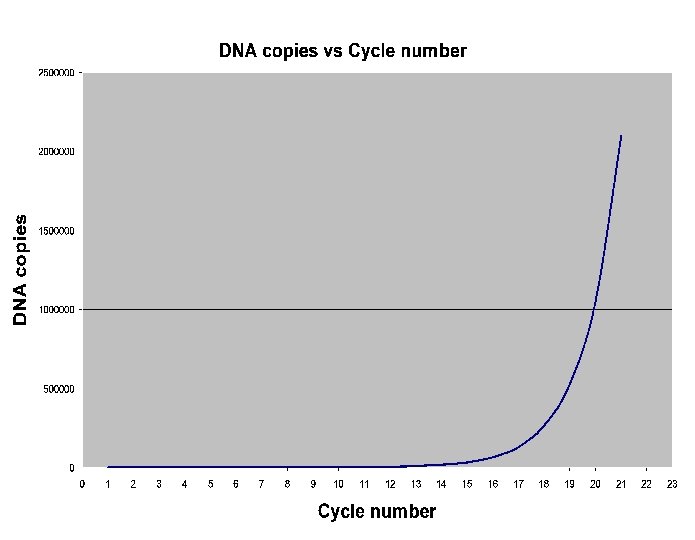
PCR: Completed Amplification Cycle n cycles will give 2 n copie.


Analyze PCR products • Check a sample by gel electrophoresis. • Is the product the size that you expected? • Is there more than one band? • Is any band the correct size? • May need to optimize the reaction conditions.

Polymerase Chain Reaction Controls • Blank reaction (Negative control reaction) – Controls for contamination – Contains all reagents except DNA template • Positive control reaction – Controls for sensitivity – Contains all reagents and a known target-containing DNA template

Procedure 1 - Prepare master Mix 2 - Program thermocycler 3 - Run the samples on thermocycler 4 - Analysis of PCR products

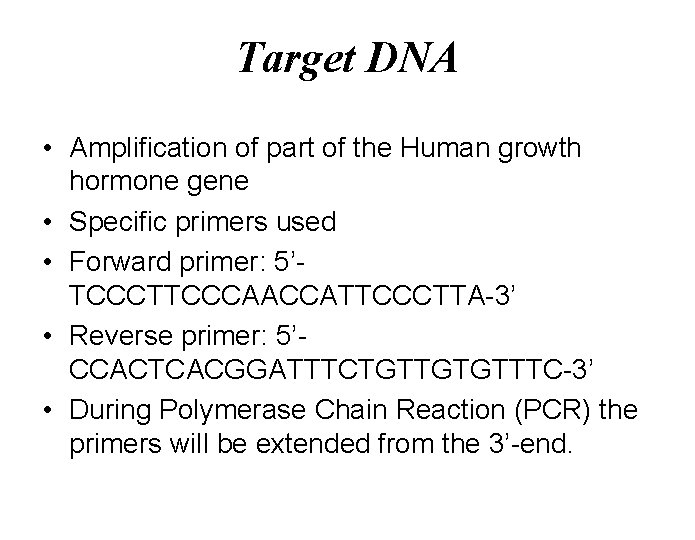
Target DNA • Amplification of part of the Human growth hormone gene • Specific primers used • Forward primer: 5’TCCCTTCCCAACCATTCCCTTA-3’ • Reverse primer: 5’CCACTCACGGATTTCTGTTGTGTTTC-3’ • During Polymerase Chain Reaction (PCR) the primers will be extended from the 3’-end.

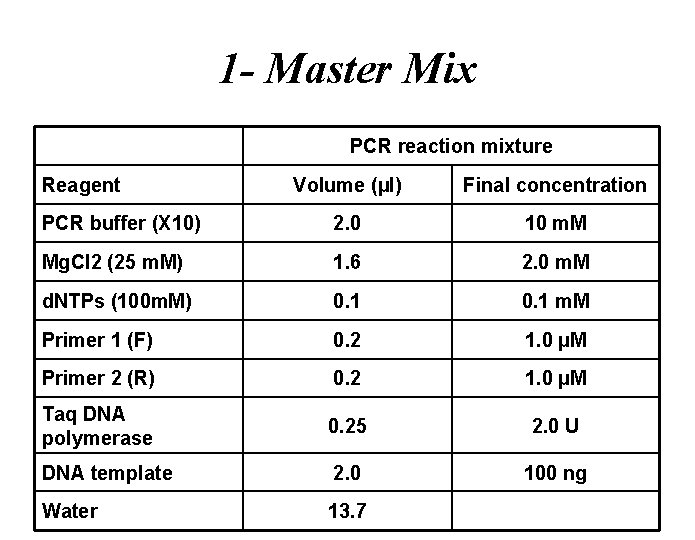
1 - Master Mix PCR reaction mixture Reagent Volume (µl) Final concentration PCR buffer (X 10) 2. 0 10 m. M Mg. Cl 2 (25 m. M) 1. 6 2. 0 m. M d. NTPs (100 m. M) 0. 1 m. M Primer 1 (F) 0. 2 1. 0 µM Primer 2 (R) 0. 2 1. 0 µM Taq DNA polymerase 0. 25 2. 0 U DNA template 2. 0 100 ng Water 13. 7

2 - Program the Thermocycler Profile is: §Step 1: Denaturation for 3 min. at 95 o. C §Step 2: 35 cycles §Melting for 60 sec. at 95 o. C §Annealing for 60 sec. at 57 o. C §Extension for 90 sec. at 72 o. C §Step 3: Final elongation for 10 min. at 72 o. C

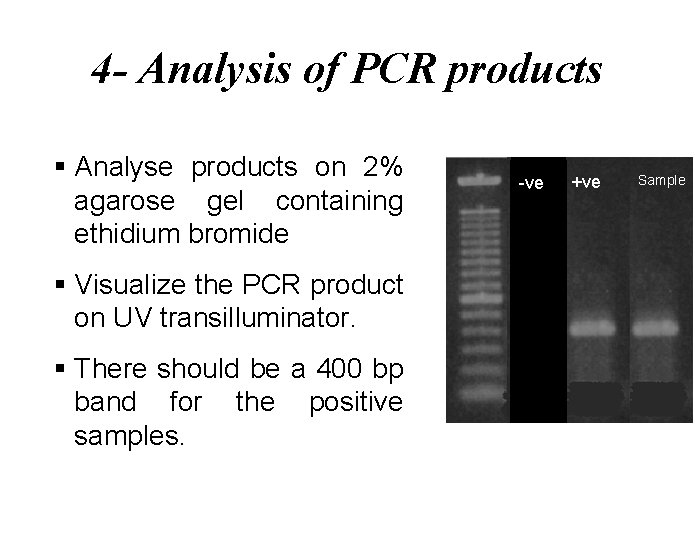
4 - Analysis of PCR products § Analyse products on 2% agarose gel containing ethidium bromide § Visualize the PCR product on UV transilluminator. § There should be a 400 bp band for the positive samples. -ve +ve Sample