The Polymerase Chain Reaction PCR By Dr NAGLAA


The Polymerase Chain Reaction (PCR) By Dr. NAGLAA FATHY Lecturer of Biochemistry & Molecular Biology Faculty of Medicine Benha University E-mail : naglaa_fathy 722000@yahoo. com Nagla. alhusseini@fmed. bu. edu. eg
What is the Polymerase Chain Reaction? It’s a means of selectively amplifying a particular segment of DNA. Ø The segment may represent a small part of a large and complex mixture of DNAs: Ø

Invented by Kary Mullis and Faloona, 1987. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Nobel Prize 1993

Kary Mullis

Did He Really Invent PCR? • The basic principle of replicating a piece of DNA using two primers had already been described by Gobind Khorana in 1971: – Kleppe et al. (1971) J. Mol. Biol. 56, 341 -346. • Progress was limited by primer synthesis and polymerase purification issues. • Mullis properly exploited amplification
PCR Specifically targets and amplifies a SINGLE sequence from within a complex mixture of DNA. Ø How is this different from cloning?

Amplify DNA PCR In vitro amplification (in a test tube) Ø Enzymatic: Taq polymerase – Temperature-resistant DNA polymerase ( Thermus aquaticus) Ø Heat resistant Ø Best for <2 kb target Ø

Takes advantage of basic requirements of replication A DNA template Ø Nucleotides Ø Primers Ø polymerase Ø PCR is DNA replication in a test tube
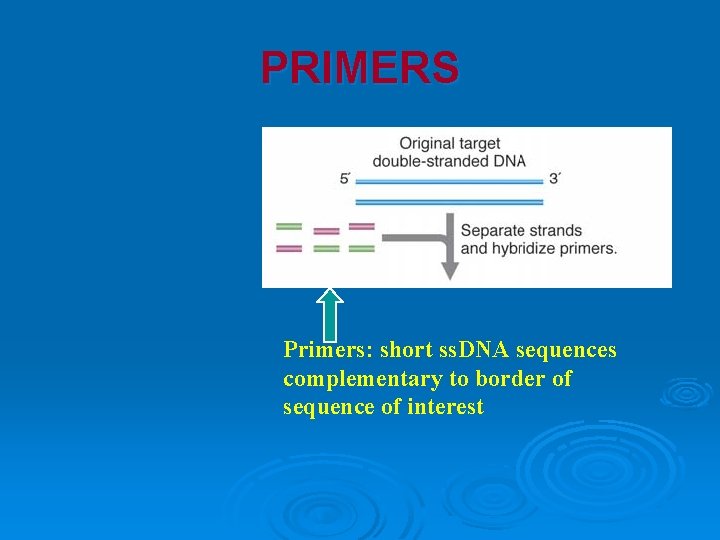
PRIMERS Primers: short ss. DNA sequences complementary to border of sequence of interest

Primers ØMust have some information about sequence flanking your target ØPrimers provide specificity
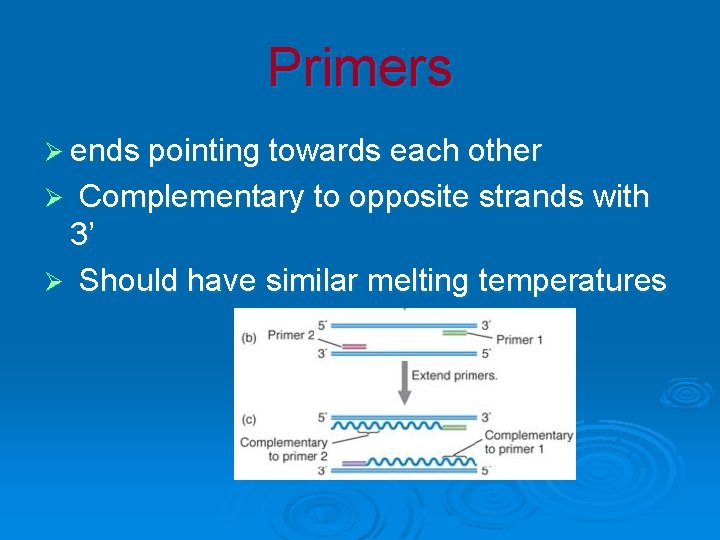
Primers Ø ends pointing towards each other Complementary to opposite strands with 3’ Ø Should have similar melting temperatures Ø

PCR Region of interest: between primers 2. Anneal 3. Extend Taq polymerase: enzymatic extension
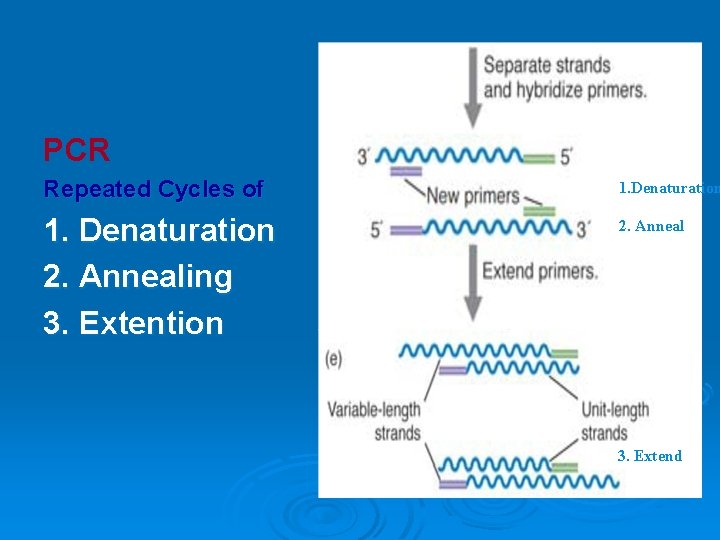
PCR Repeated Cycles of 1. Denaturation 2. Annealing 3. Extention 2. Anneal 3. Extend

Melting temperature Tmo. C : Temperature at which half possible H bonds are formed. Tmo. C = 2(A+T) + 4(G+C) 5/ - AGACTCAGAGAGAACCC-3/ 4 Gs 5 Cs 7 As 1 T Tmo. C= (4 x 9) + (2 x 8) = 36+16 = 520 C Annealing T =Tm 0 C -5

Heat-stable polymerase is vital to the ease of the process…

Ø Thermus aquaticus from hot springs in Yellowstone National Park, USA.
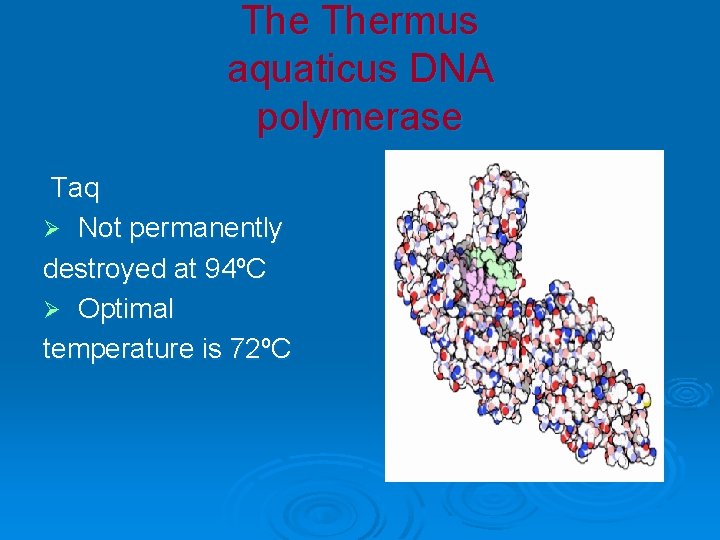
The Thermus aquaticus DNA polymerase Taq Ø Not permanently destroyed at 94ºC Ø Optimal temperature is 72ºC

Problems with Taq DNA polymerase - thermostable Ø Lack of 3′-5′ exonuclease – proofreading ►Error rate = 2 × 10 -4 nucleotdes/cycle Ø Newer polymerases have high fidelity High fidelity polymerase - Hi. Fi Taq
Termplates for PCR Ø Small amount of template In theory a single molecule Ø Do not need to isolate sequence of interest Ø DNA template need not be highly purified Ø DNA is stable in absence of nucleases Ø

Templates for PCR �� Dried blood �� Semen stains �� Vaginal swabs �� Single hair �� Finger nail scrapings �� Egyptian mummies �� Buccal Swab �� Tooth brushes

Basic reaction ►Thermocycling, PCR machine �� Previously – need to overlay oil to prevent evaporation �� Automatically Change temperature �� Temperature gradient
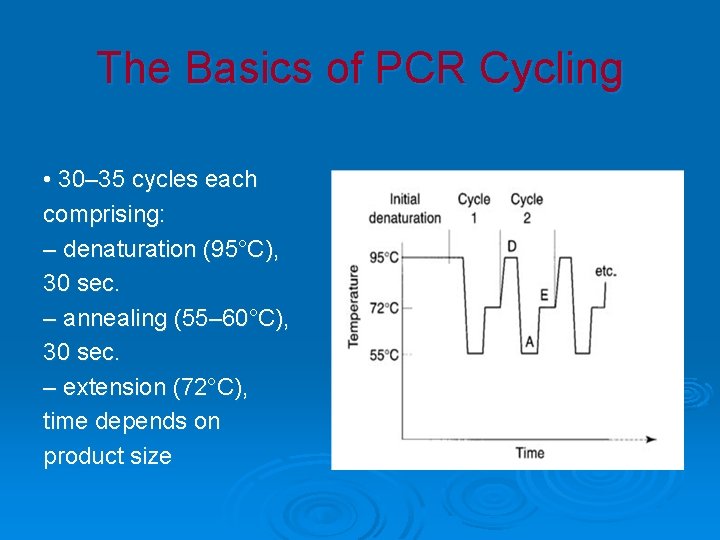
The Basics of PCR Cycling • 30– 35 cycles each comprising: – denaturation (95°C), 30 sec. – annealing (55– 60°C), 30 sec. – extension (72°C), time depends on product size

How many copies? • No target products are made until the third cycle. • The accumulation is not strictly a doubling at each cycle in the early phase. • At 30 cycles there are 1, 073, 741, 764 target copies (~1× 109).
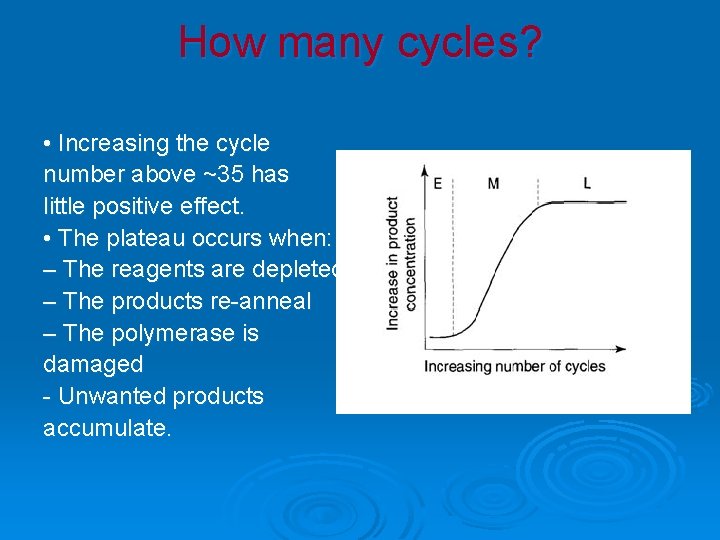
How many cycles? • Increasing the cycle number above ~35 has little positive effect. • The plateau occurs when: – The reagents are depleted – The products re-anneal – The polymerase is damaged - Unwanted products accumulate.
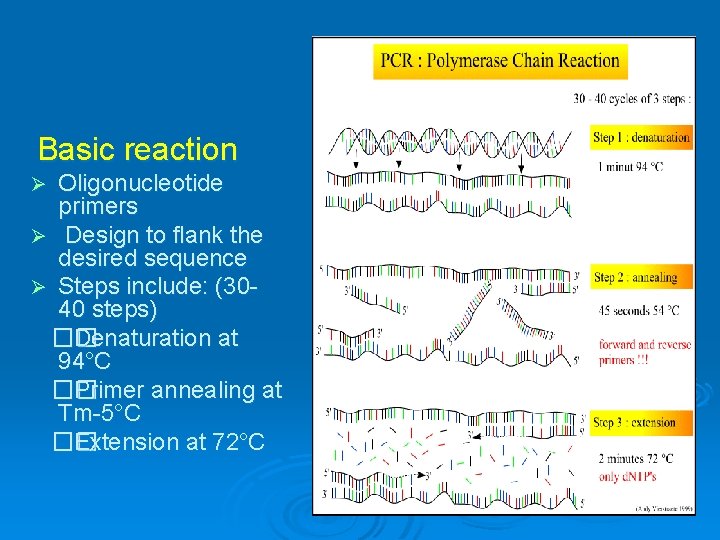
Basic reaction Oligonucleotide primers Ø Design to flank the desired sequence Ø Steps include: (3040 steps) �� Denaturation at 94°C �� Primer annealing at Tm-5°C �� Extension at 72°C Ø

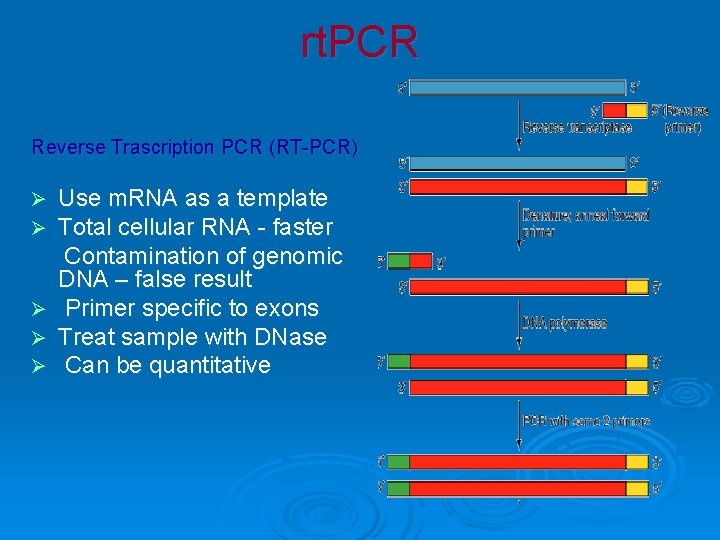
rt. PCR Reverse Trascription PCR (RT-PCR) Ø Ø Ø Use m. RNA as a template Total cellular RNA - faster Contamination of genomic DNA – false result Primer specific to exons Treat sample with DNase Can be quantitative

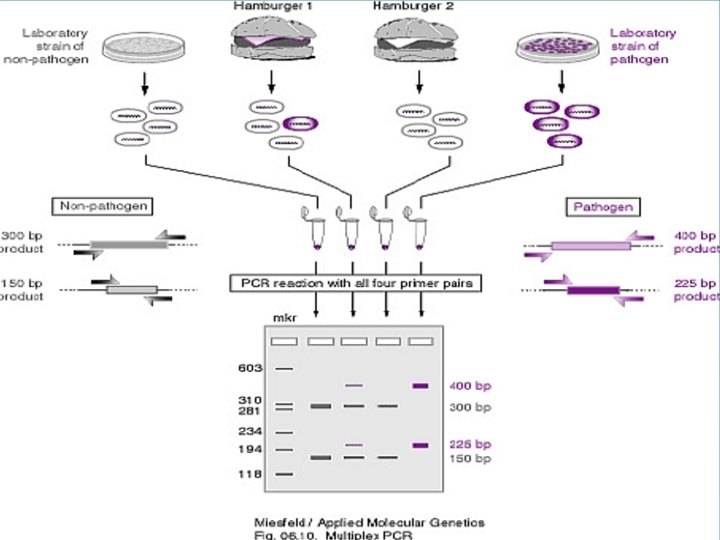
Multiplex PCR Simultaneously modification of more than one locus in the same reaction Ø Rapid and convenient – screening Ø Included different set of primers Ø

Quantitative or Real Time PCR Monitors the fluorescence emitted during the reaction as an indicator of amplicon production Ø during each PCR cycle. �� The parameter CT (threshold cycle) is defined as the cycle number at which the fluorescence emission exceeds the fixed threshold (background). Ø
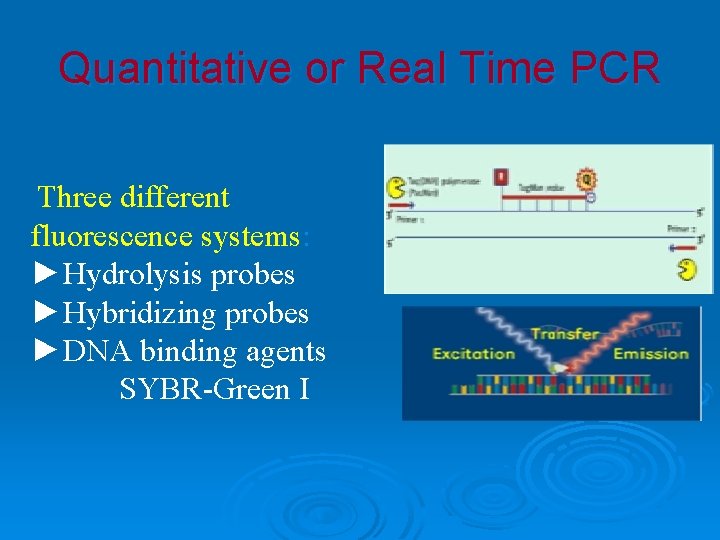
Quantitative or Real Time PCR Three different fluorescence systems: ►Hydrolysis probes ►Hybridizing probes ►DNA binding agents SYBR-Green I
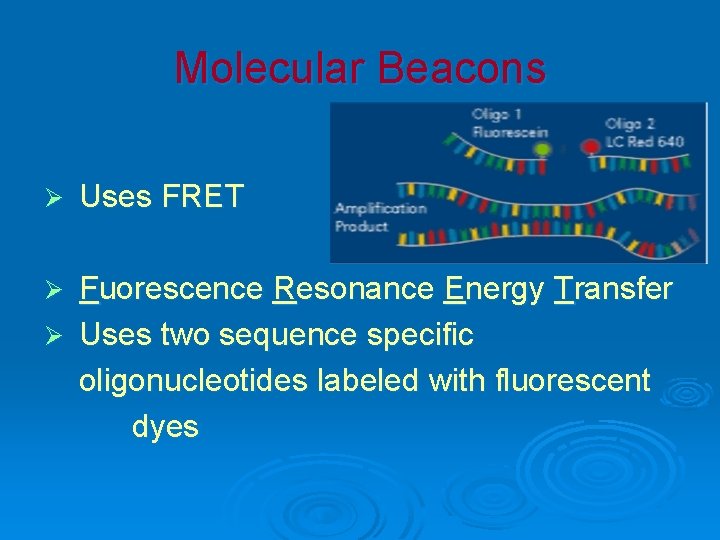
Molecular Beacons Ø Uses FRET Fuorescence Resonance Energy Transfer Ø Uses two sequence specific oligonucleotides labeled with fluorescent dyes Ø
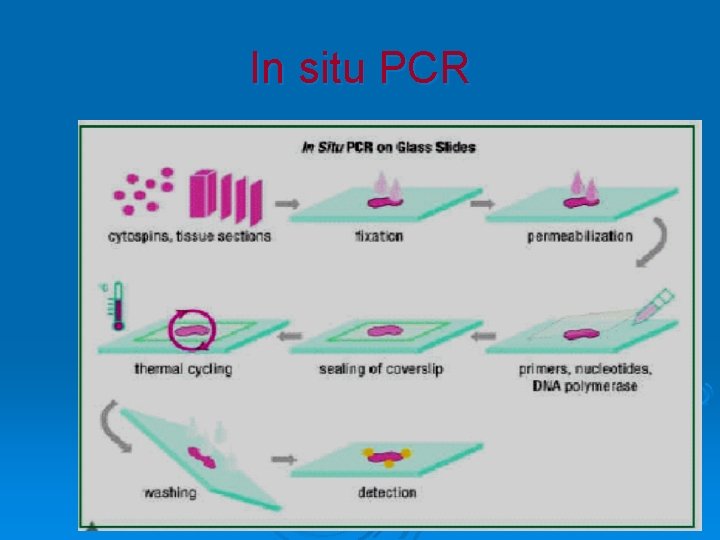
In situ PCR
Applications of PCR Mutation detection. Ø Diagnosis or screening of acquired diseases, : e. g. AIDS, HBV & HCV. Ø Prenatal diagnosis Ø DNA profiling in forensic science Ø Quantitation of m. RNA in cells or tissues. Ø
Problems with PCR Ø Contamination Theoretically one molecule can amplify Ø Takes one mismatch early on to amplify the wrong fragment Ø

Dr. Naglaa Fathy
- Slides: 37