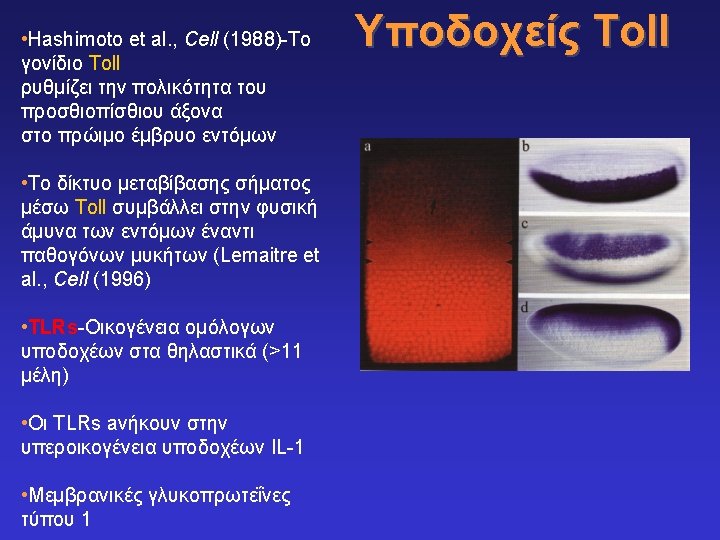
The Pattern recognition hypothesis Charlie Janeway 1989 With





















- Slides: 65




The Pattern recognition hypothesis, Charlie Janeway 1989 • ‘’With brilliant insight, Janeway predicted in 1989 that pattern recognition receptors would mediate the body’s ability to recognize invasion by microorganisms. This striking prediction was made first on theoretical grounds and subsequently incisive experimental work in his laboratory established the underlying mechanisms. In this way, Janeway was one of the key fathers of what has become the new field of innate immunity, perhaps the most exciting area of immunologic research in recent times. “ Yale School of Medicine Dean David Kessler,

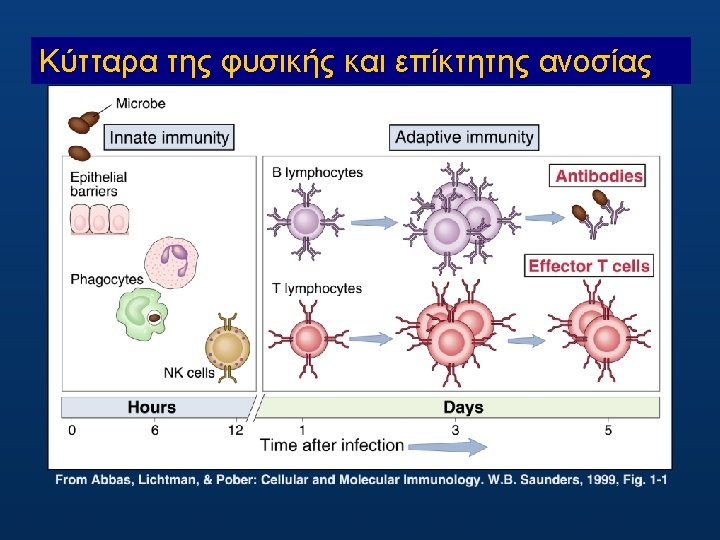
Δυσδιάκριτα τα όρια ανάμεσα στη φυσική και επίκτητη ανοσοαπόκριση? Nature Reviews Immunology 9, 302 -303 (30 April 2009) ORIGINAL RESEARCH PAPER: Sun, J. C. , Beilke, J. N. & Lanier, L. L. Adaptive immune features of n a t u r a l k i l l e r c e l l s. N a t u r e, 1 1 J a n 2 0 0 9










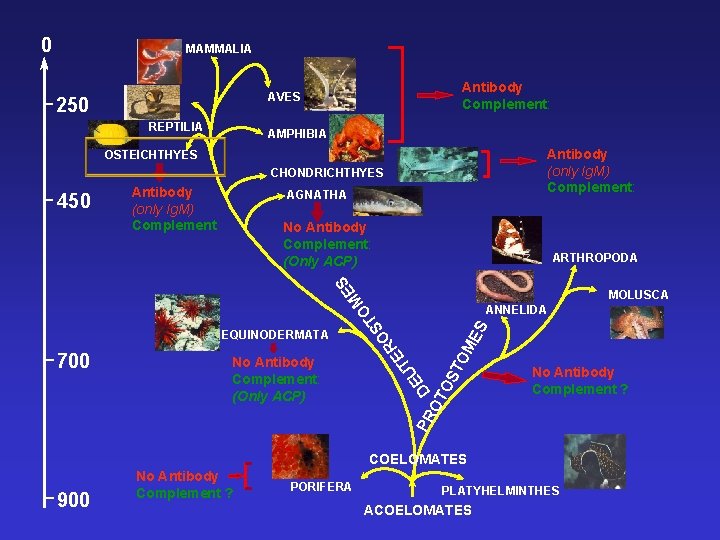
0 MAMMALIA Antibody Complement: AVES 250 REPTILIA AMPHIBIA Antibody (only Ig. M) Complement: OSTEICHTHYES CHONDRICHTHYES Antibody (only Ig. M) Complement AGNATHA No Antibody Complement: (Only ACP) ARTHROPODA ES 450 ES OM PR D OT OS T No Antibody Complement: (Only ACP) T U E 700 ANNELIDA O R E EQUINODERMATA O ST M MOLUSCA No Antibody Complement ? COELOMATES 900 No Antibody Complement ? PORIFERA PLATYHELMINTHES ACOELOMATES




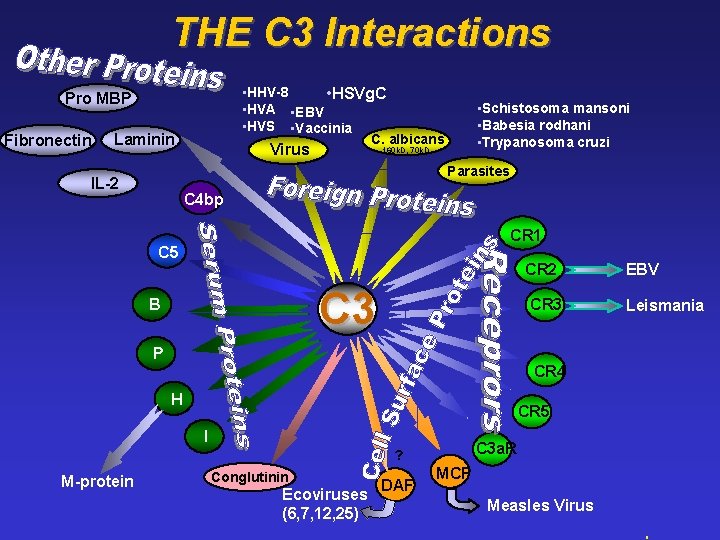
THE C 3 Interactions • HHV-8 • HSVg. C • HVA • EBV • HVS • Vaccinia C. albicans Pro MBP Fibronectin Laminin Virus • Schistosoma mansoni • Babesia rodhani • Trypanosoma cruzi 160 k. D, 70 k. D Parasites IL-2 C 4 bp CR 1 C 5 CR 2 C 3 B CR 3 P CR 4 H CR 5 I C 3 a. R ? M-protein Conglutinin Ecoviruses (6, 7, 12, 25) DAF MCP Measles Virus EBV Leismania









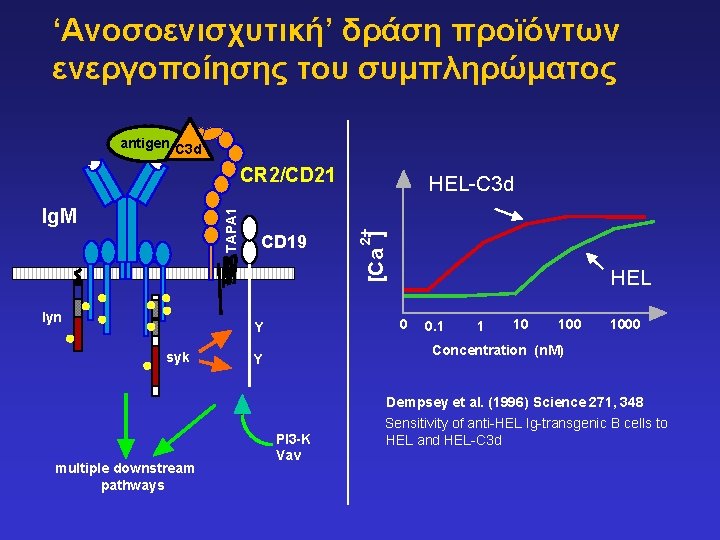
‘Ανοσοενισχυτική’ δράση προϊόντων ενεργοποίησης του συμπληρώματος antigen C 3 d CR 2/CD 21 lyn HEL 0 Y syk 2+ CD 19 [Ca ] TAPA 1 Ig. M HEL-C 3 d 0. 1 1 10 1000 Concentration (n. M) Y Dempsey et al. (1996) Science 271, 348 multiple downstream pathways PI 3 -K Vav Sensitivity of anti-HEL Ig-transgenic B cells to HEL and HEL-C 3 d

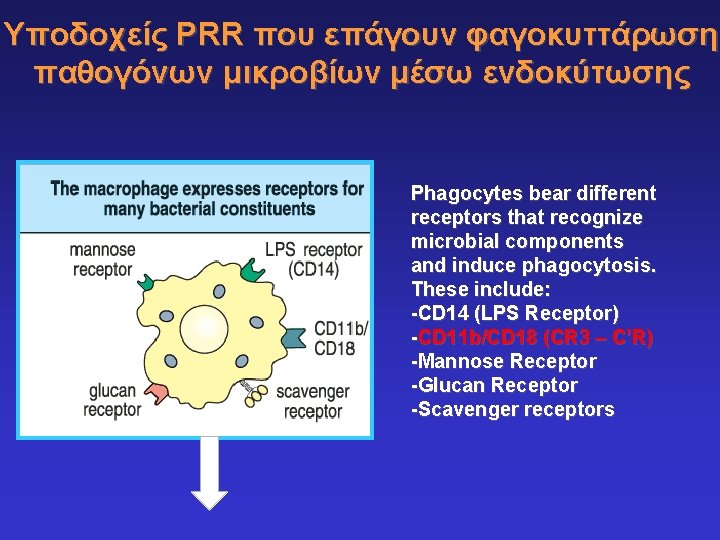
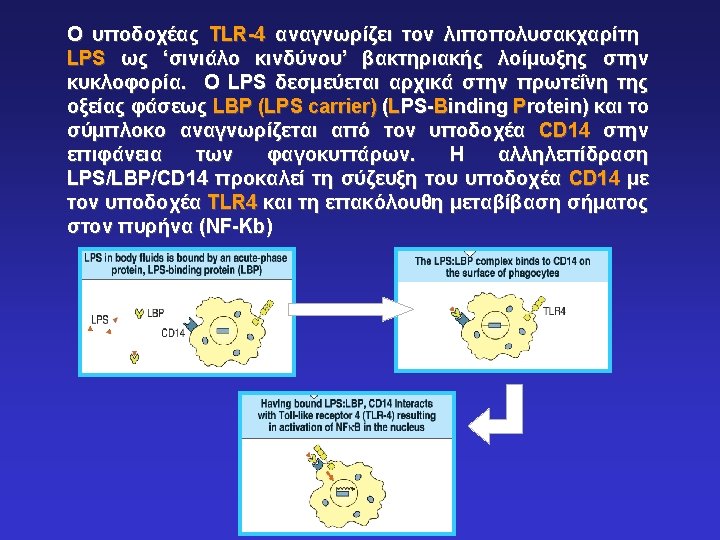
Υποδοχείς PRR που επάγουν φαγοκυττάρωση παθογόνων μικροβίων μέσω ενδοκύτωσης Phagocytes bear different receptors that recognize microbial components and induce phagocytosis. These include: -CD 14 (LPS Receptor) -CD 11 b/CD 18 (CR 3 – C’R) -Mannose Receptor -Glucan Receptor -Scavenger receptors

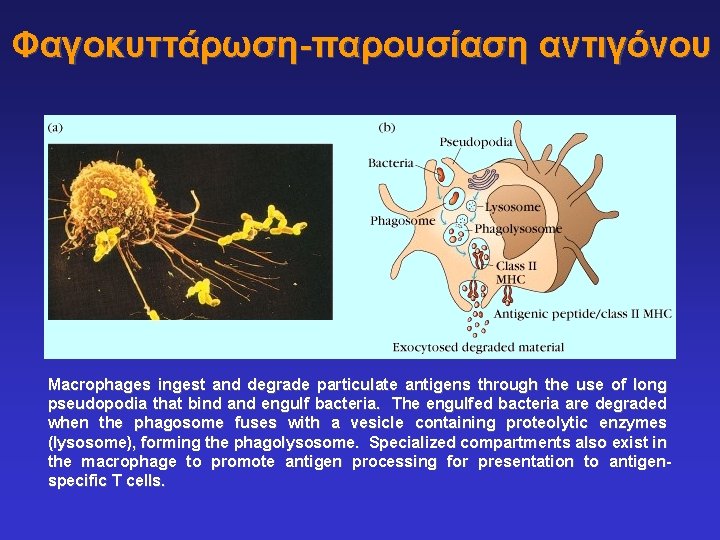
Φαγοκυττάρωση-παρουσίαση αντιγόνου Macrophages ingest and degrade particulate antigens through the use of long pseudopodia that bind and engulf bacteria. The engulfed bacteria are degraded when the phagosome fuses with a vesicle containing proteolytic enzymes (lysosome), forming the phagolysosome. Specialized compartments also exist in the macrophage to promote antigen processing for presentation to antigenspecific T cells.



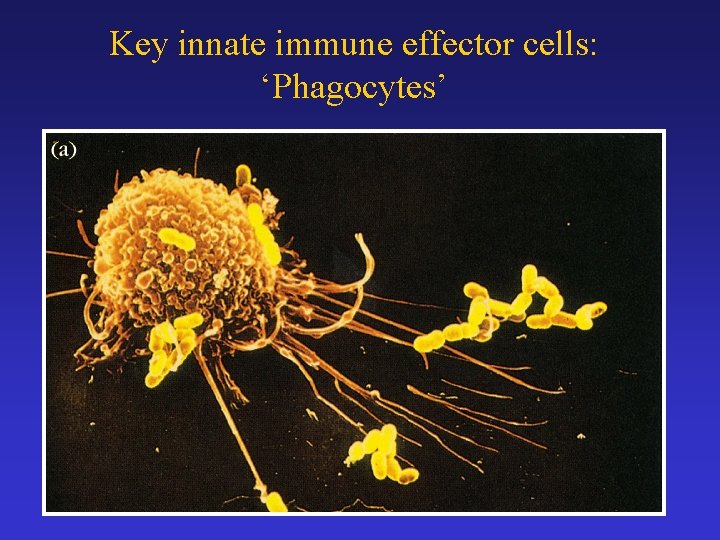
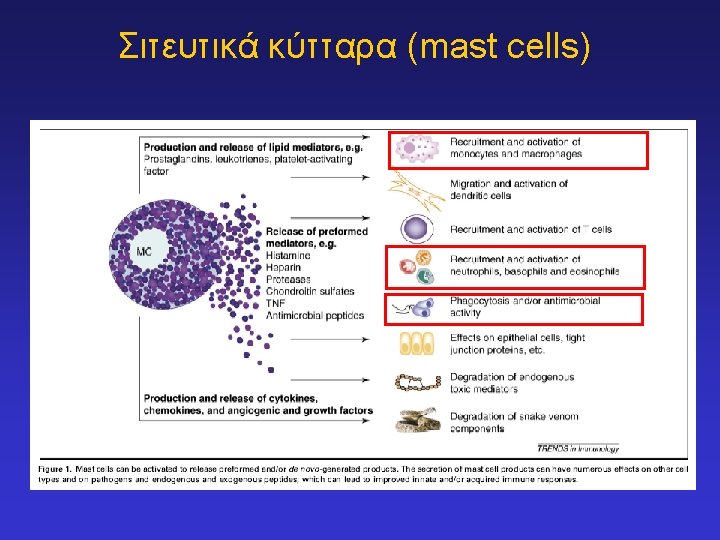
Key innate immune effector cells: ‘Phagocytes’

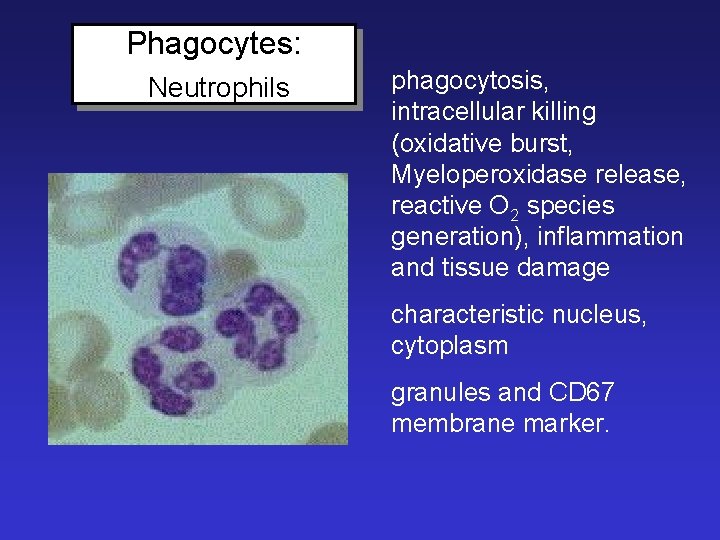
Phagocytes: Neutrophils phagocytosis, intracellular killing (oxidative burst, Myeloperoxidase release, reactive O 2 species generation), inflammation and tissue damage characteristic nucleus, cytoplasm granules and CD 67 membrane marker.

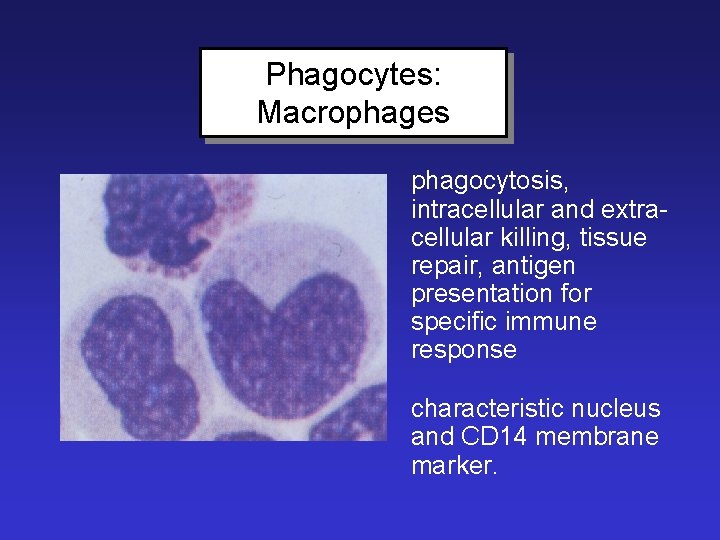
Phagocytes: Macrophages phagocytosis, intracellular and extracellular killing, tissue repair, antigen presentation for specific immune response characteristic nucleus and CD 14 membrane marker.

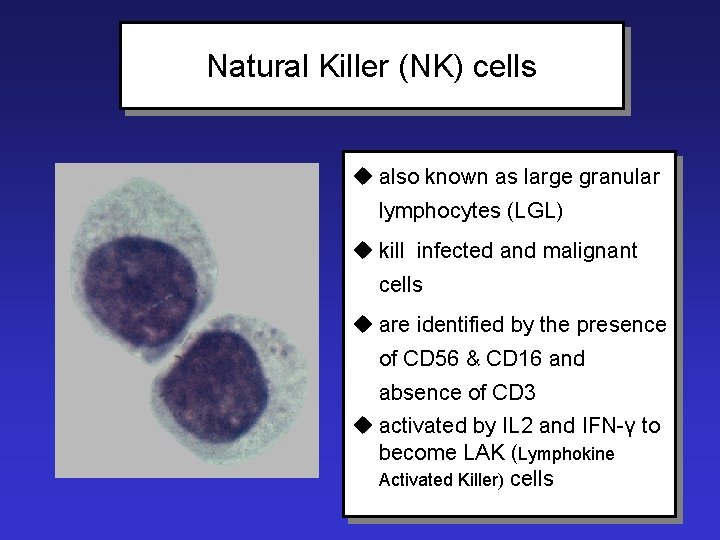
Natural Killer (NK) cells u also known as large granular lymphocytes (LGL) u kill infected and malignant cells u are identified by the presence of CD 56 & CD 16 and absence of CD 3 u activated by IL 2 and IFN-γ to become LAK (Lymphokine Activated Killer) cells

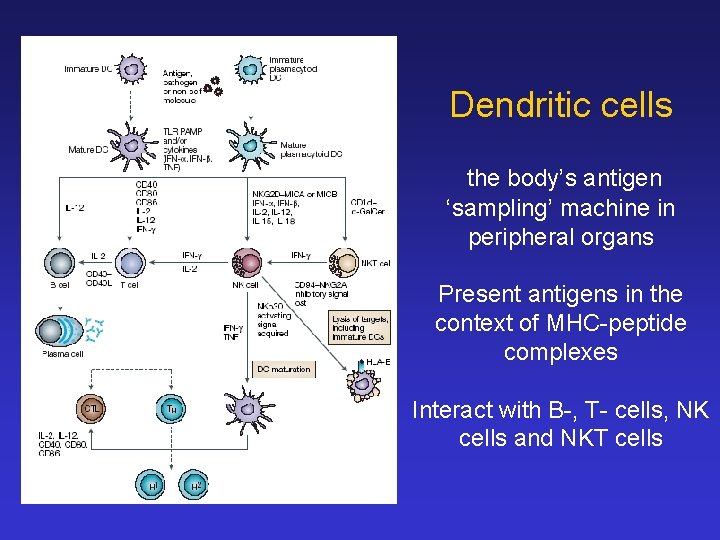
Dendritic cells the body’s antigen ‘sampling’ machine in peripheral organs Present antigens in the context of MHC-peptide complexes Interact with B-, T- cells, NK cells and NKT cells

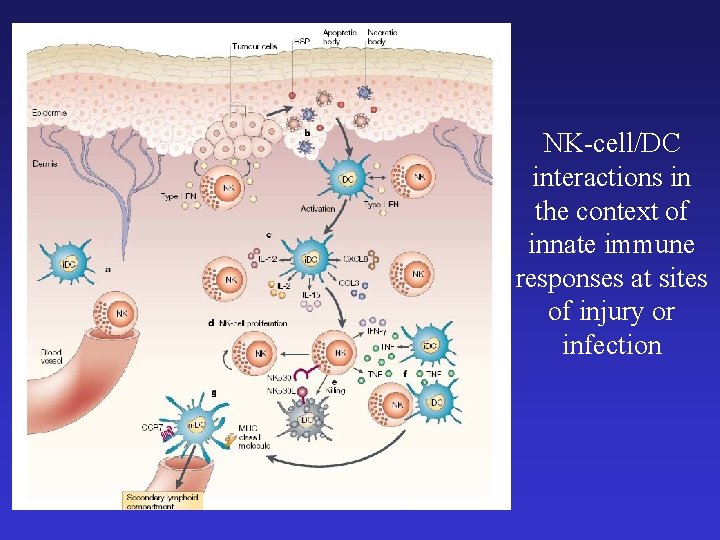
NK-cell/DC interactions in the context of innate immune responses at sites of injury or infection





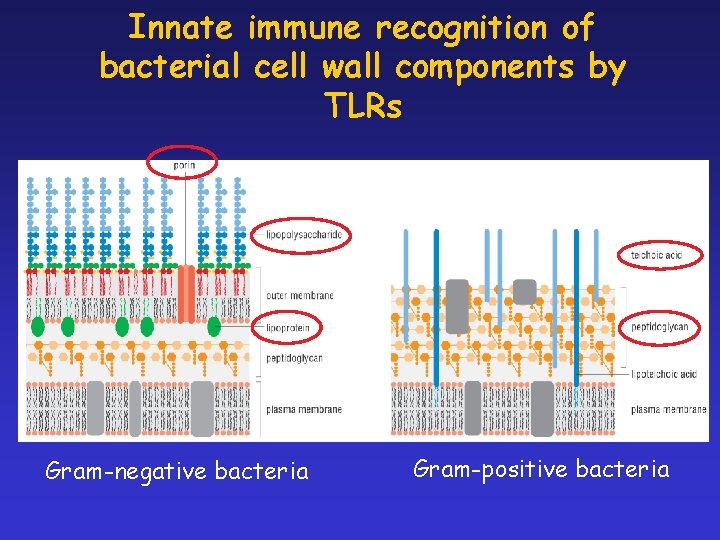
Innate immune recognition of bacterial cell wall components by TLRs Gram-negative bacteria Gram-positive bacteria

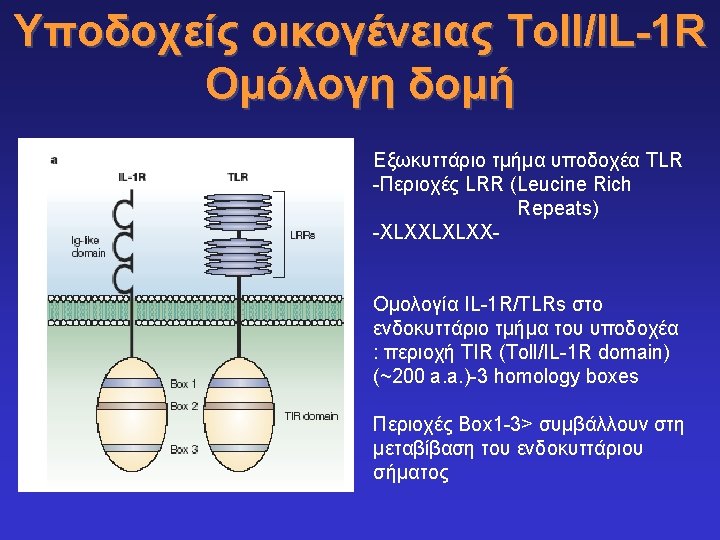
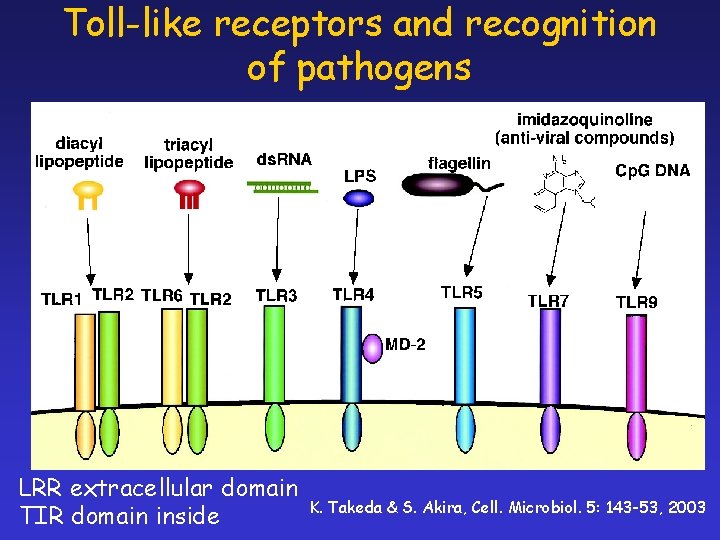
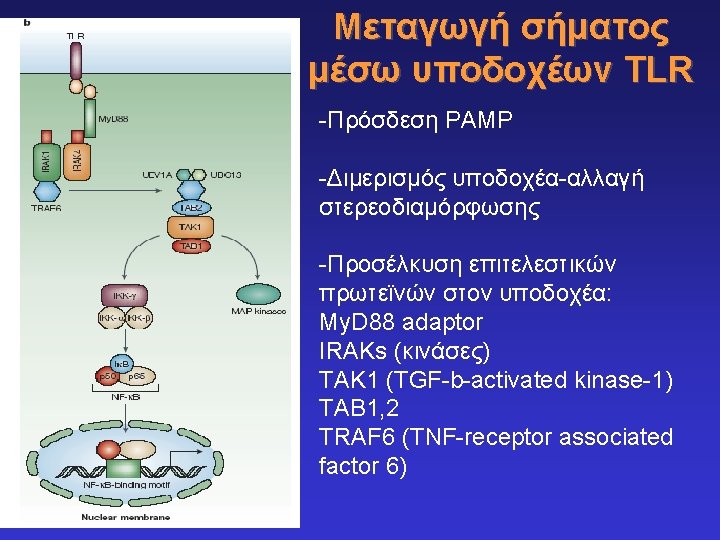
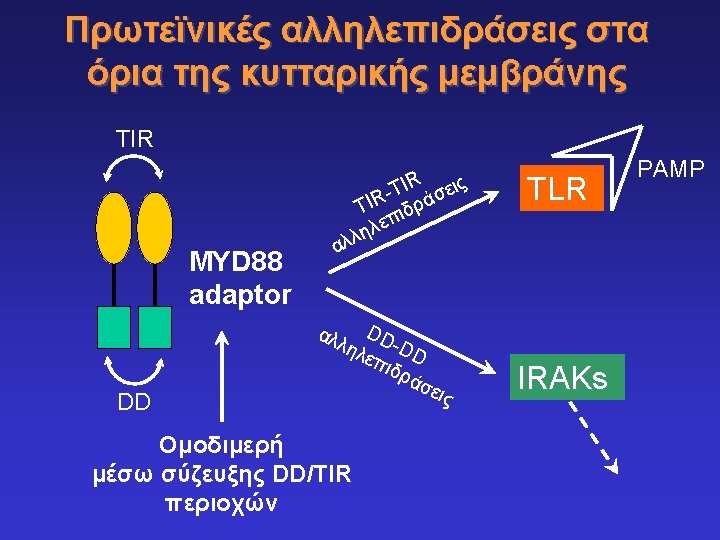
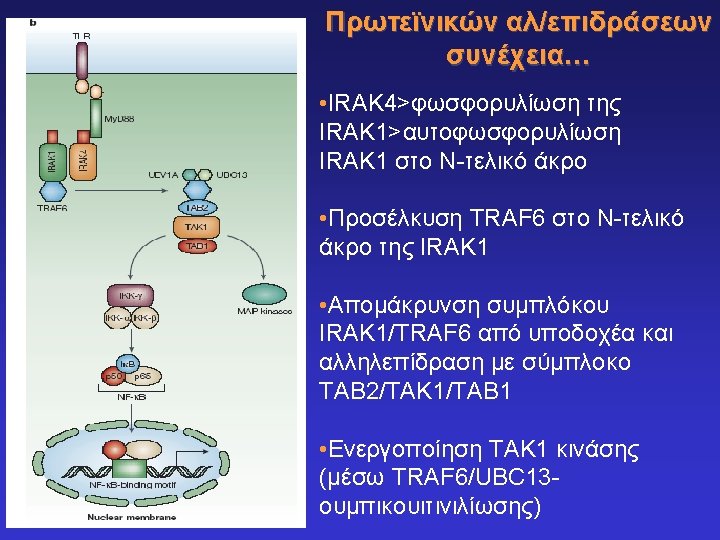
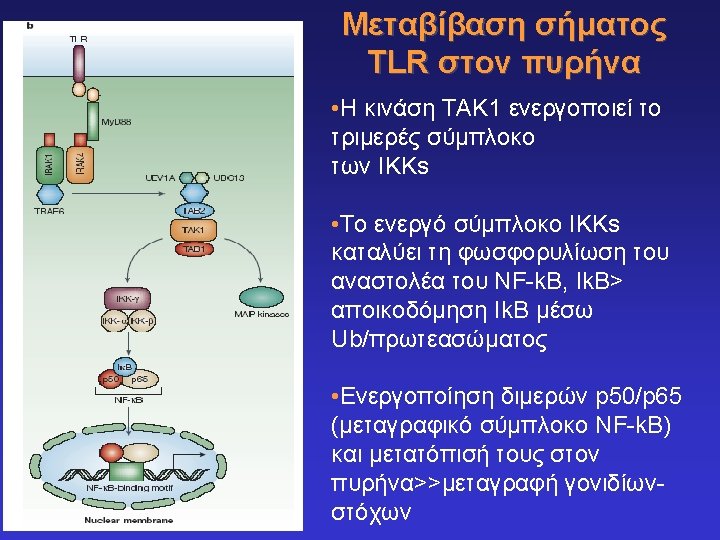
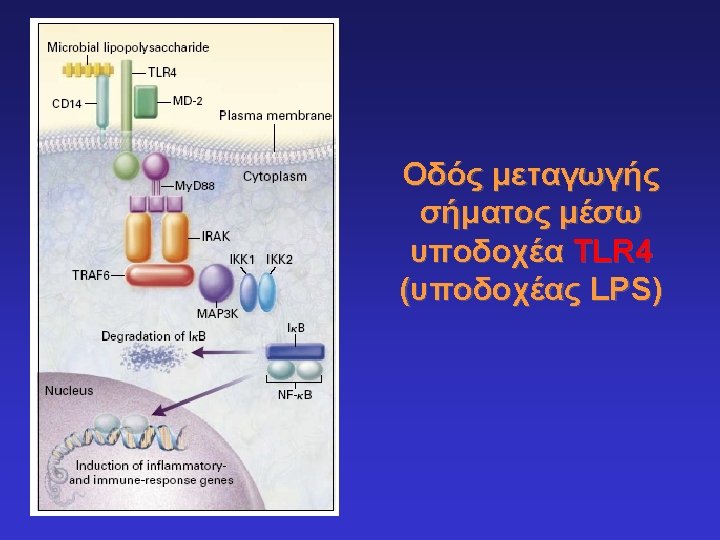
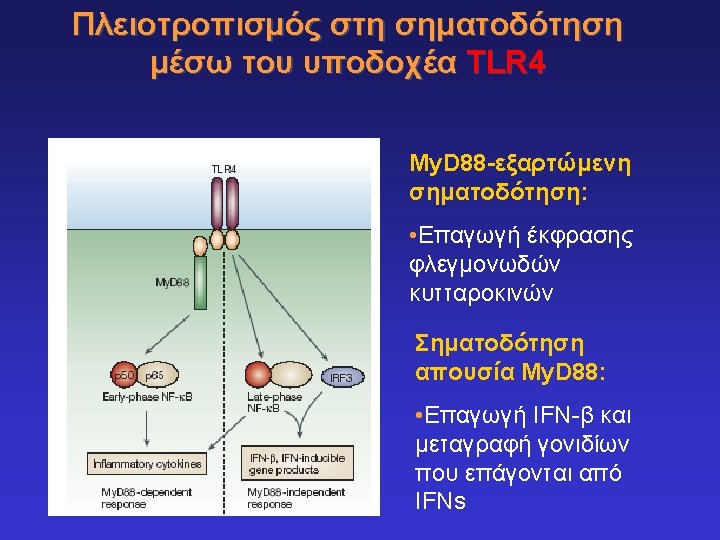
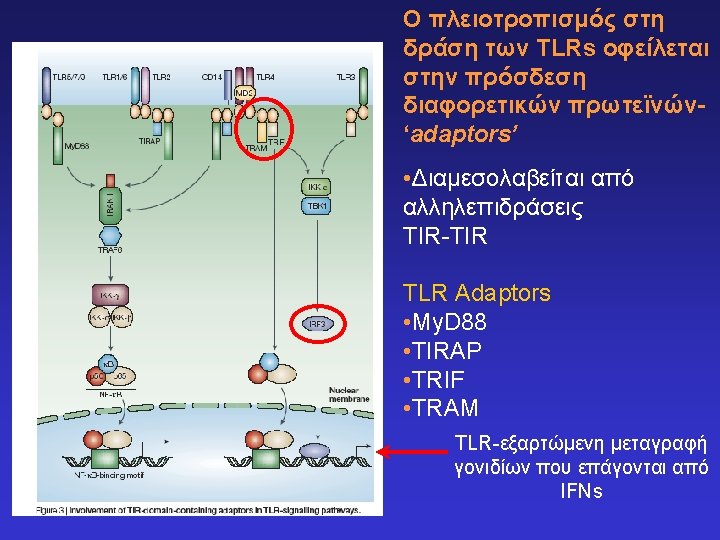
Toll-like receptors and recognition of pathogens LRR extracellular domain TIR domain inside K. Takeda & S. Akira, Cell. Microbiol. 5: 143 -53, 2003

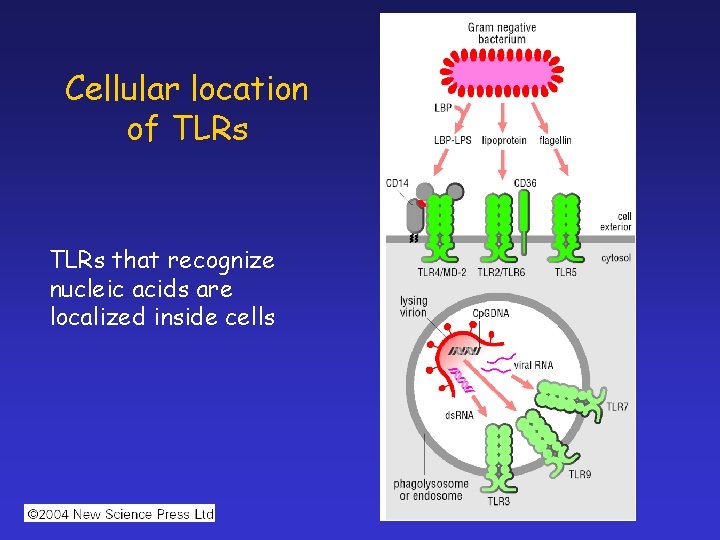
Cellular location of TLRs that recognize nucleic acids are localized inside cells

















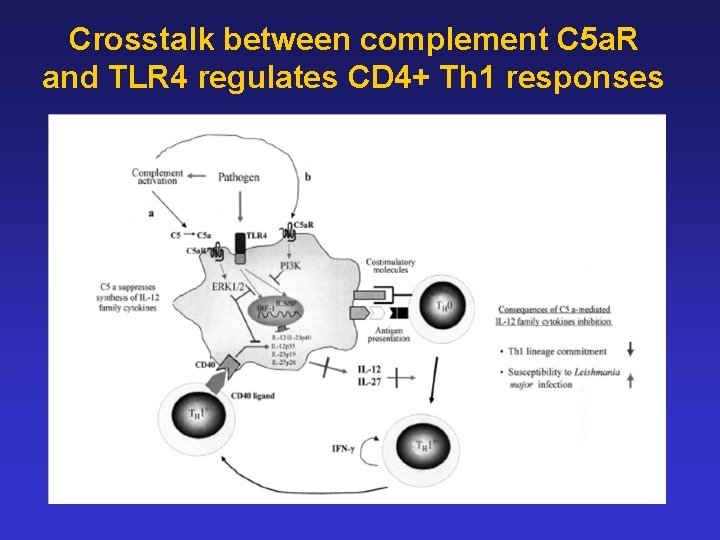
Crosstalk between complement C 5 a. R and TLR 4 regulates CD 4+ Th 1 responses

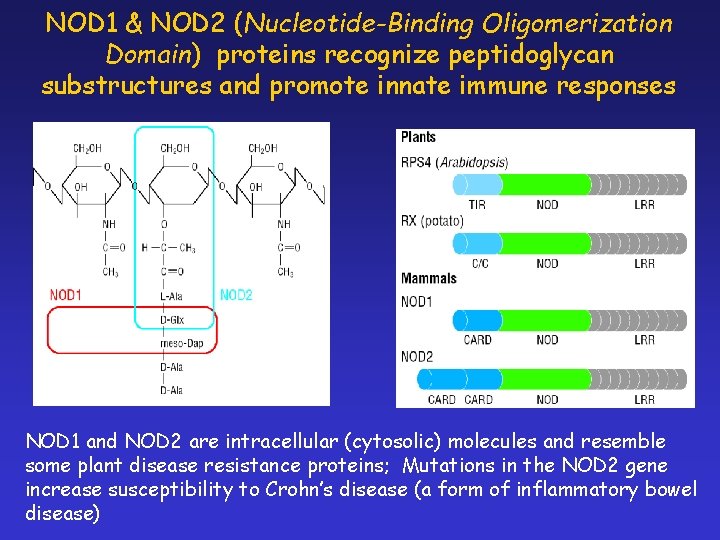
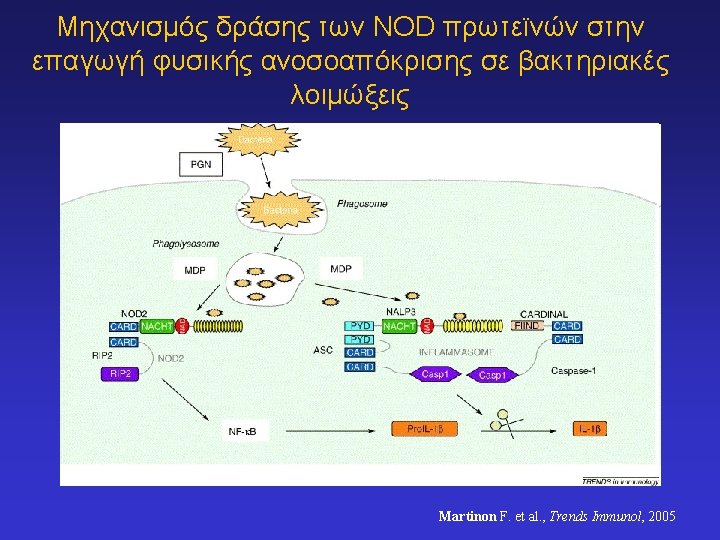
NOD 1 & NOD 2 (Nucleotide-Binding Oligomerization Domain) proteins recognize peptidoglycan substructures and promote innate immune responses NOD 1 and NOD 2 are intracellular (cytosolic) molecules and resemble some plant disease resistance proteins; Mutations in the NOD 2 gene increase susceptibility to Crohn’s disease (a form of inflammatory bowel disease)


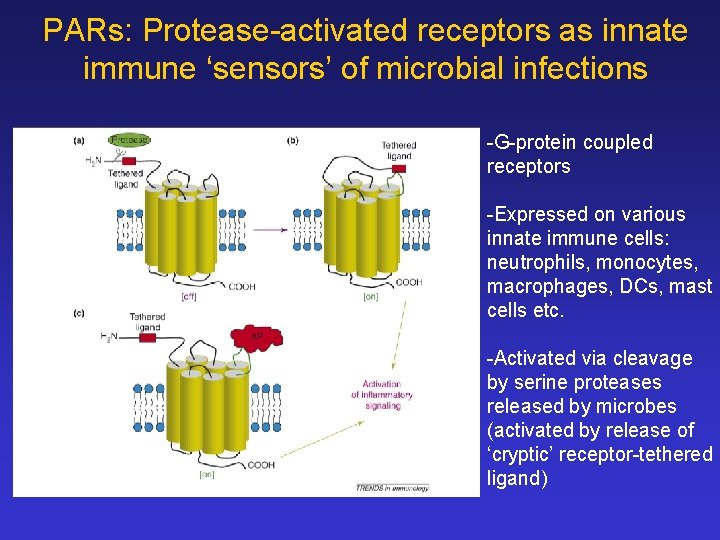
PARs: Protease-activated receptors as innate immune ‘sensors’ of microbial infections -G-protein coupled receptors -Expressed on various innate immune cells: neutrophils, monocytes, macrophages, DCs, mast cells etc. -Activated via cleavage by serine proteases released by microbes (activated by release of ‘cryptic’ receptor-tethered ligand)