The Pathway Tools Software and Bio Cyc Database

The Pathway Tools Software and Bio. Cyc Database Collection Peter D. Karp, Ph. D. Bioinformatics Research Group SRI International pkarp@ai. sri. com http: //www. ai. sri. com/pkarp/talks/ Bio. Cyc. org Eco. Cyc. org, Meta. Cyc. org, Human. Cyc. org 1 SRI International Bioinformatics
Use Cases for Pathway Tools and Bio. Cyc l Development of organism-specific DBs (modelorganism DBs) that span many biological datatypes l Web publishing of those DBs with a powerful set of query and visualization tools l Computational inferences of metabolic pathways, pathway hole fillers, operons, transport reactions l Visual tools for analysis of omics data l Tools for analysis of biological networks l Comparative analysis tools l Metabolic engineering l Bio. Cyc is a Web portal for genome and pathway information 2 SRI International Bioinformatics
Bio. Cyc Collection of 673 Pathway/Genome Databases l. Pathway/Genome Database (PGDB) – combines information about l Pathways, reactions, substrates l Enzymes, transporters l Genes, replicons l Transcription factors/sites, promoters, operons l. Tier l l 1: Literature-Derived PGDBs Meta. Cyc Eco. Cyc -- Escherichia coli K-12 l. Tier 2: Computationally-derived DBs, Some Curation -- 28 PGDBs l Human. Cyc l Mycobacterium tuberculosis l. Tier 3: Computationally-derived DBs, No Curation -- 643 DBs 3 SRI International Bioinformatics

Pathway Tools Software l Patho. Logic l Predicts operons, metabolic network, pathway hole fillers, from genome l Computational creation of new Pathway/Genome Databases l Pathway/Genome Editors l Distributed curation of PGDBs l Distributed object database system, interactive editing tools l Pathway/Genome Navigator l WWW publishing of PGDBs l Querying, visualization of pathways, chromosomes, operons l Analysis operations u u Pathway visualization of gene-expression data Global comparisons of metabolic networks Briefings in Bioinformatics 11: 40 -79 2010 4 SRI International Bioinformatics
Obtaining a PGDB for Organism of Interest l Find existing curated PGDB l Find existing PGDB in Bio. Cyc l Create your own l Curated pathway DBs now exist for most biomedical model organisms 5 SRI International Bioinformatics

Pathway Tools Software: PGDBs Created Outside SRI l 2, 100+ licensees: 180 groups applying software to 1, 600 organisms l. Saccharomyces cerevisiae, SGD project, Stanford University l 135 pathways / 565 publications l. Candida albicans, CGD project, Stanford University ldicty. Base, Northwestern University l. Mouse, MGD, Jackson Laboratory l. Drosophila, Fly. Base, Harvard University l. Under development: l C. elegans, Worm. Base l. Arabidopsis thaliana, TAIR, Carnegie Institution of Washington l 288 pathways / 2282 publications l. Plant. Cyc, Carnegie Institution of Washington l. Six Solanaceae species, Cornell University l. Gramene. DB, Cold Spring Harbor Laboratory l. Medicago truncatula, Samuel Roberts Noble Foundation 6 SRI International Bioinformatics
Meta. Cyc: Metabolic Encyclopedia l Describe a representative sample of every experimentally determined metabolic pathway l Describe properties of metabolic enzymes l Literature-based DB with extensive references and commentary l Meta. Cyc now assigns more than twice as many reactions to pathways as does KEGG Nucleic Acids Research 2010 7 SRI International Bioinformatics
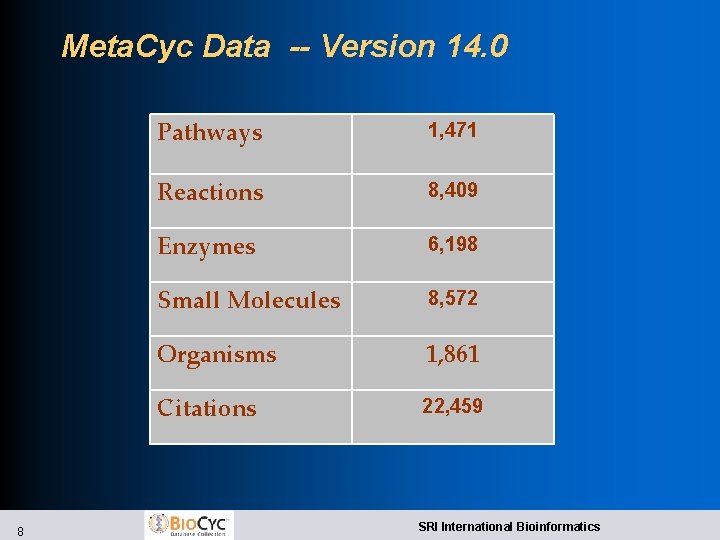
Meta. Cyc Data -- Version 14. 0 8 Pathways 1, 471 Reactions 8, 409 Enzymes 6, 198 Small Molecules 8, 572 Organisms 1, 861 Citations 22, 459 SRI International Bioinformatics

Pathway Tools Survey Publication l Karp 10 et al, Briefings in Bioinformatics 2010 11: 40 -79. SRI International Bioinformatics
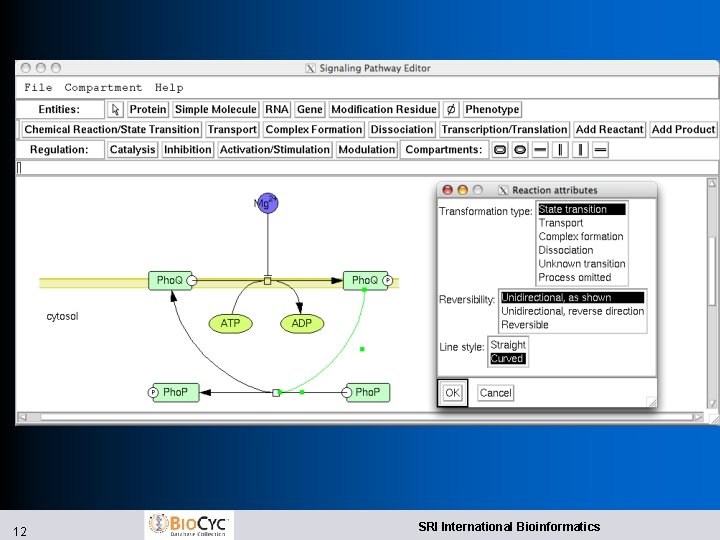
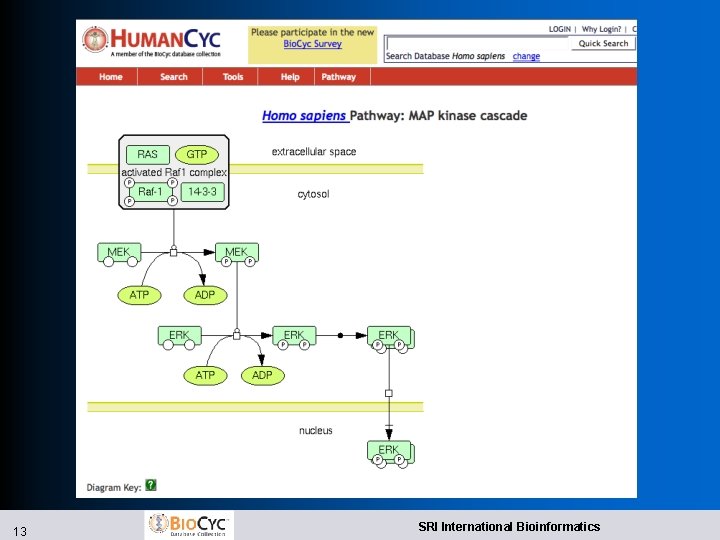
Signaling Pathway Editor l Signaling pathways use different visual conventions than metabolic pathways l Look and feel based of our tool based on Cell. Designer, SBGN l Manual l 11 layout Can’t yet be included in Cellular Overview Diagram SRI International Bioinformatics
12 SRI International Bioinformatics
13 SRI International Bioinformatics

Improved Web Overviews l Implemented using Open. Layers l Zoomable, draggable, searchable, paintable l Cellular Overview l Highlight compounds, reactions, enzymes, genes by name, substring, with autocomplete l Highlight genes from file l Superimpose omics data l Regulatory Overview l Draw connections between a gene and its regulators, regulatees l Show full diagram or only highlighted genes 14 SRI International Bioinformatics
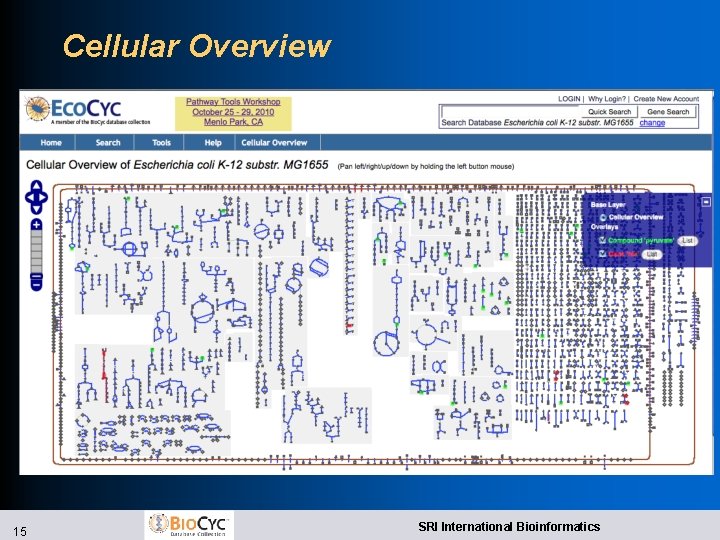
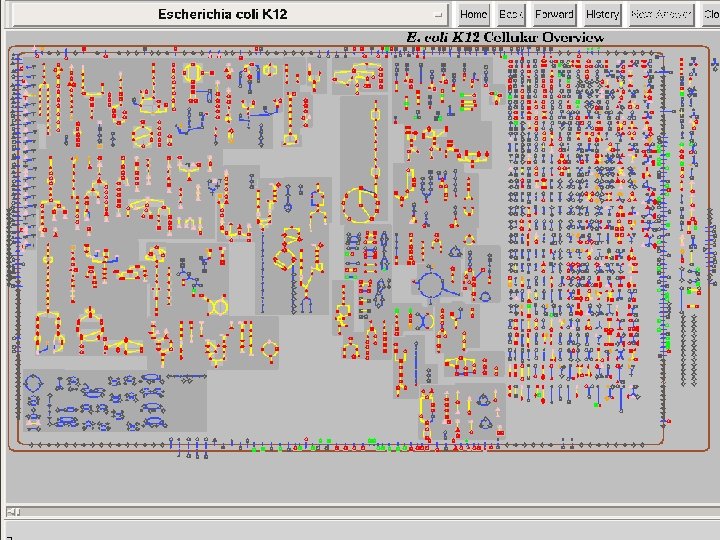
Cellular Overview 15 SRI International Bioinformatics
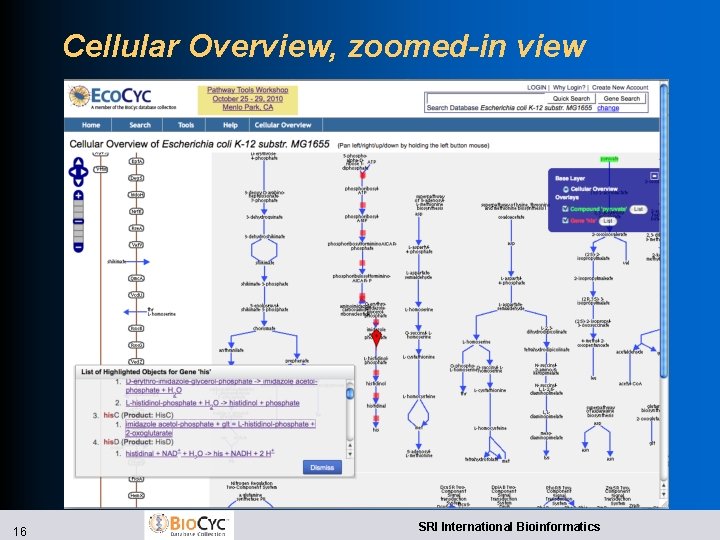
Cellular Overview, zoomed-in view 16 SRI International Bioinformatics
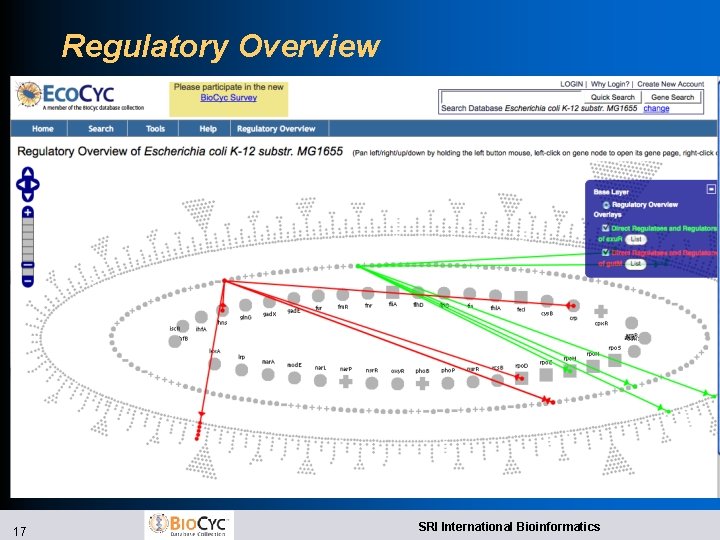
Regulatory Overview 17 SRI International Bioinformatics
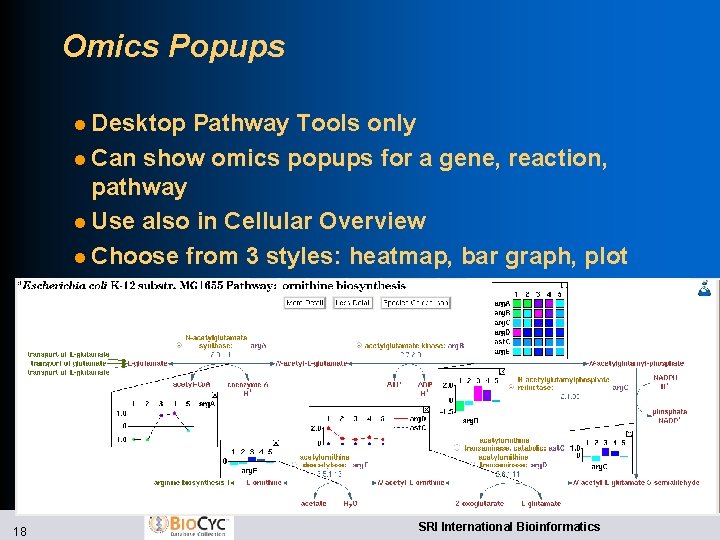
Omics Popups l Desktop Pathway Tools only l Can show omics popups for a gene, reaction, pathway l Use also in Cellular Overview l Choose from 3 styles: heatmap, bar graph, plot 18 SRI International Bioinformatics
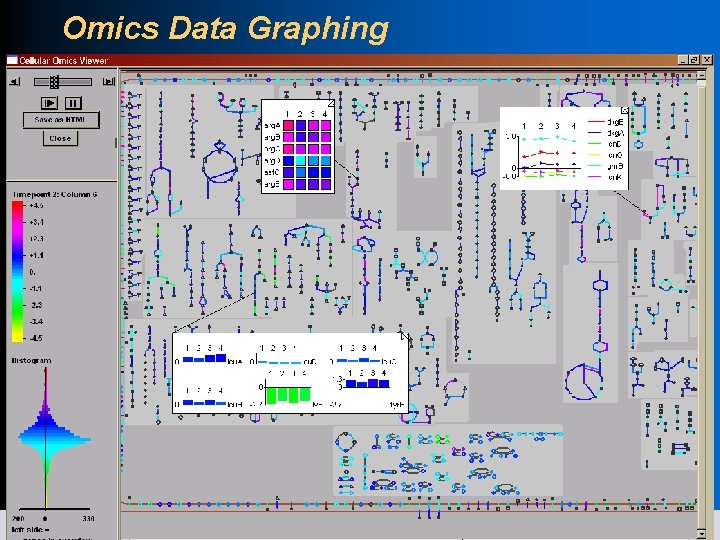
Omics Data Graphing 19 SRI International Bioinformatics
Pathway Tools Captures All Bacterial Regulation Mechanisms l Regulation of transcription l By transcription factors l By attenuation l Regulation of translation l By proteins and small RNAs l Regulation of protein activity l By covalent modification (e. g. , phosphorylation) l By non-covalent modification (e. g. , allosteric inhibitors) l Support: 20 Schema, editing tools, display tools SRI International Bioinformatics
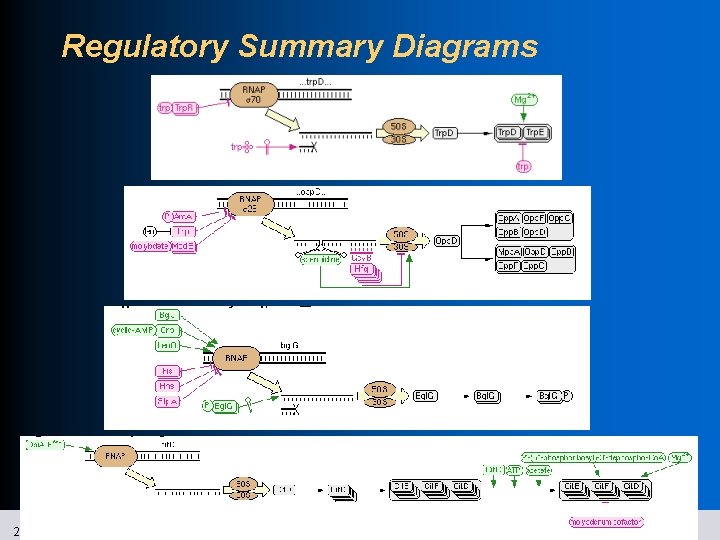
Regulatory Summary Diagrams 21 SRI International Bioinformatics

Other Recent Enhancements l Phases I and II of upgrade to Pathway Tools Web mode l Phase III still to come l Ability l 22 to customize pathway displays via Web site Pathway Customize SRI International Bioinformatics

Reachability Analysis of Metabolic Networks l l l Given: l A PGDB for an organism l A set of initial metabolites Infer: l What set of products can be synthesized by the smallmolecule metabolism of the organism Motivations: l Quality control for PGDBs u Verify that a known growth medium yields known essential compounds Experiment with other growth media l Experiment with reaction knock-outs Limitations l Cannot properly handle compounds required for their own synthesis l Nutrients needed for reachability may be a superset of those required for growth l l Romero and Karp, Pacific Symposium on Biocomputing, 2001 23 SRI International Bioinformatics
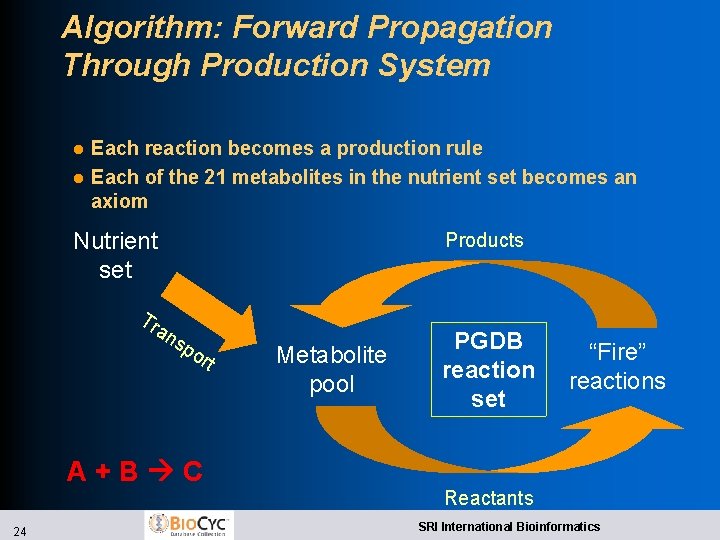
Algorithm: Forward Propagation Through Production System l l Each reaction becomes a production rule Each of the 21 metabolites in the nutrient set becomes an axiom Nutrient set Tr an Products sp o rt A+B C 24 Metabolite pool PGDB reaction set “Fire” reactions Reactants SRI International Bioinformatics
25 SRI International Bioinformatics

Coming Soon l Bio. Cyc / Eco. Cyc / Human. Cyc will support Web services for data retrieval l i. Phone app for Bio. Cyc / Eco. Cyc / Human. Cyc and other PGDBs 26 SRI International Bioinformatics

Acknowledgements l. SRI l l. Funding Suzanne Paley, Ron Caspi, Ingrid Keseler, Carol Fulcher, Markus Krummenacker, Alex Shearer, Tomer Altman, Joe Dale, Fred Gilham, Pallavi Kaipa l l. Eco. Cyc l Collaborators Julio Collado-Vides, Robert Gunsalus, Ian Paulsen l sources: NIH National Institute of General Medical Sciences NIH National Center for Research Resources l. Meta. Cyc Bio. Cyc. org Collaborators l Sue Rhee, Peifen Zhang, Kate Bio. Cyc Dreherwebinars: biocyc. org/webinar. shtml Learn more from l Lukas Mueller, Anuradha SRI International Bioinformatics 27
- Slides: 26