The overlapping community structure of complex networks Introduction


















- Slides: 41

The overlapping community structure of complex networks

Introduction • • Networks and complex systems The structure of networks Finding communities Devisive and agglomerative methods Network construction in examples Statistical features The importance of observing networks

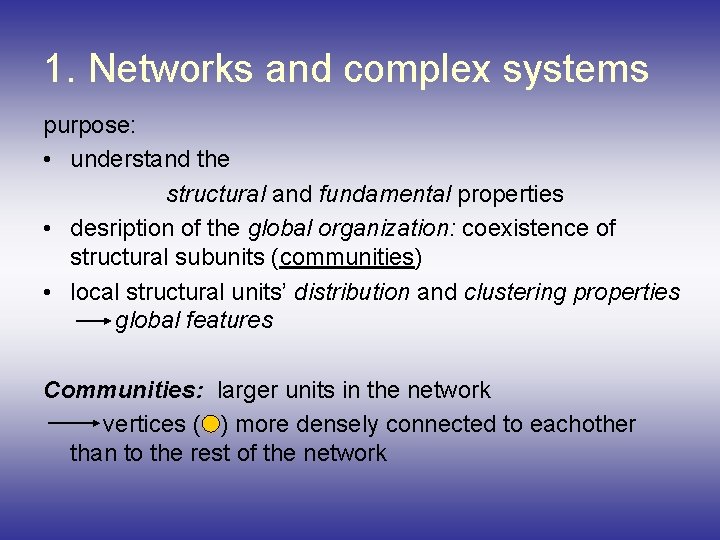
1. Networks and complex systems purpose: • understand the structural and fundamental properties • desription of the global organization: coexistence of structural subunits (communities) • local structural units’ distribution and clustering properties global features Communities: larger units in the network vertices ( ) more densely connected to eachother than to the rest of the network

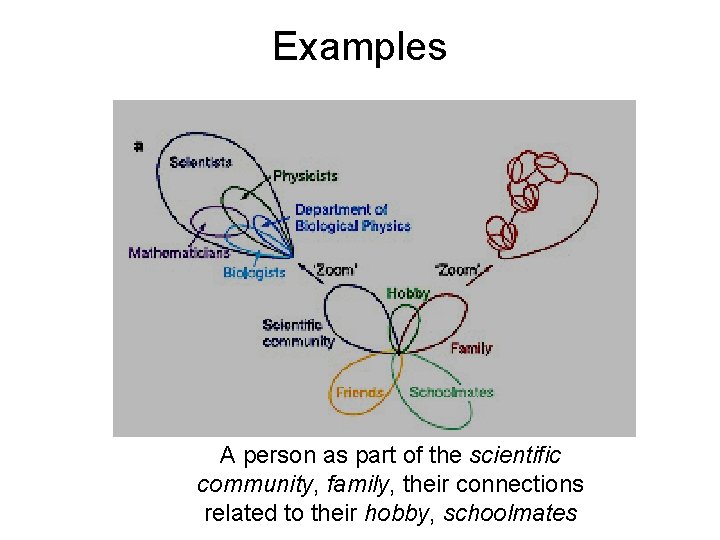
Examples A person as part of the scientific community, family, their connections related to their hobby, schoolmates

such blocks: • in the industrial sectors • functionally related proteins • word association communities (next illustration)

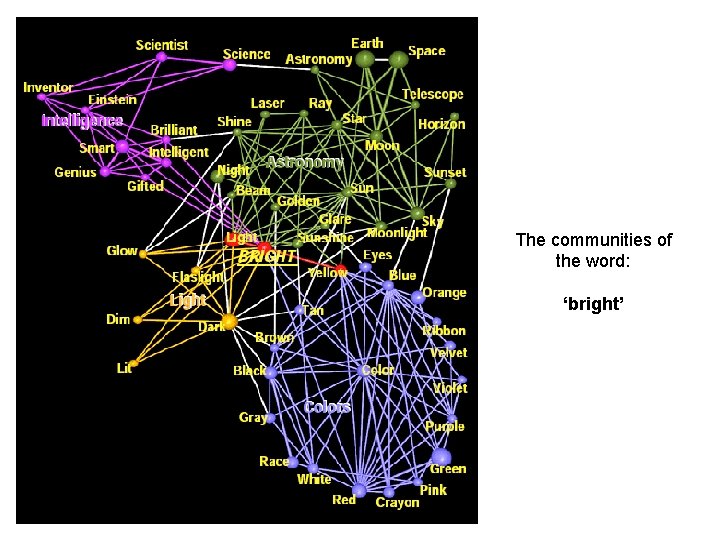
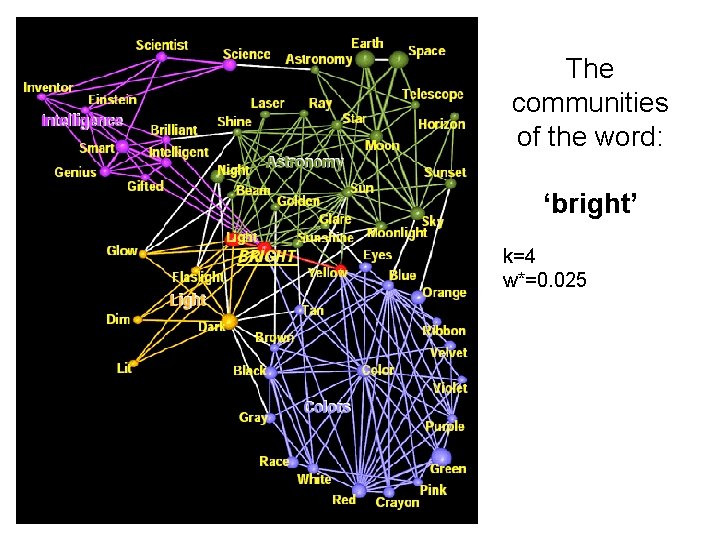
The communities of the word: ‘bright’

Problems with the identifications of communities • different kind of methods: à usually they don’t allow for overlapping communities However overlapping is important. à devide networks into smaller peaces

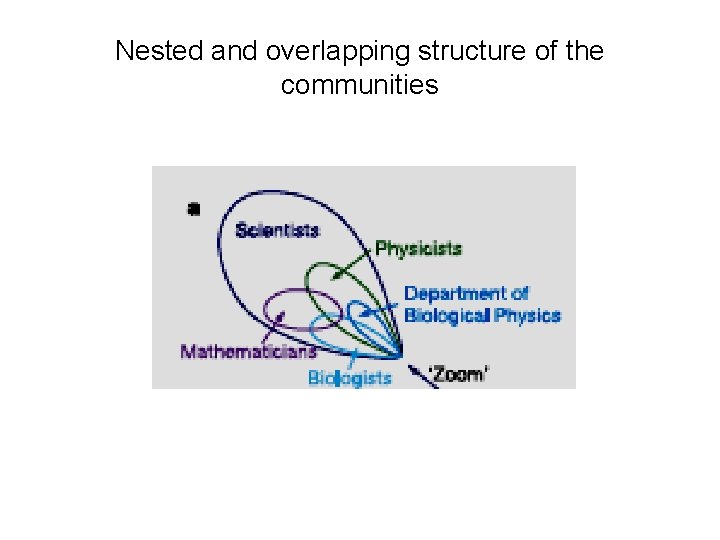
Nested and overlapping structure of the communities

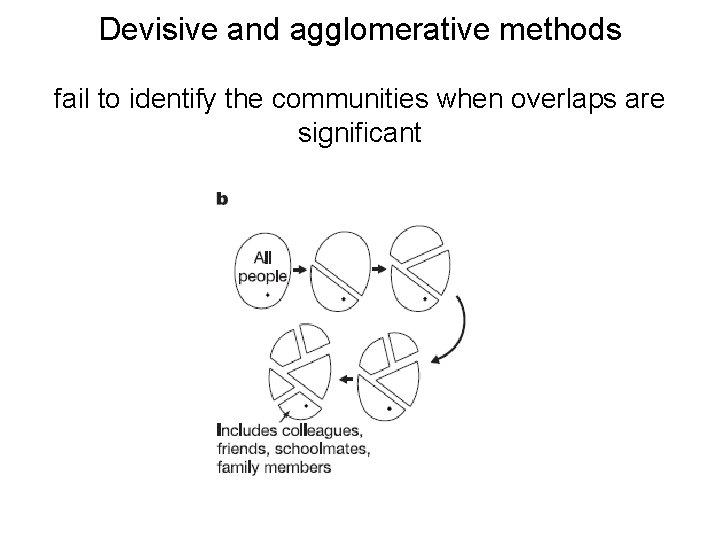
Devisive and agglomerative methods fail to identify the communities when overlaps are significant

• We would like to discuss an approach to analysing the main statistical features we need new characteristic quantities • Introduce a technique for exploring overlapping communities on a large scale

2. The stucture of networks • Clusters/communities: Those parts of the network in which the nodes are more highly connected to each other than to the rest of the network. • Membership number: mi number of communities that node i belongs to • Overlap size between α and β communities: Sovα, β the number of nodes which communities α and β share

• Community degree: dαcom the number of those links which are overlaps • Size of community α: sαcom number of nodes We would like to examine the distribution of these quantities: m sov dcom scom → → P(m) P(sov) P(dcom) P(scom)

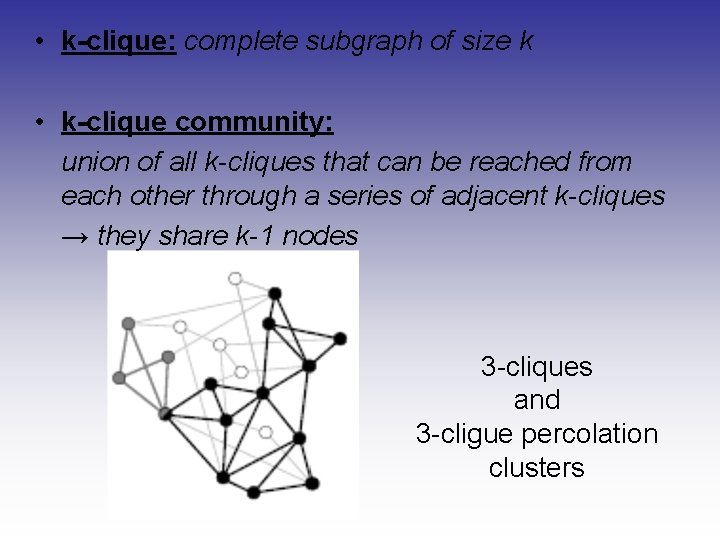
• k-clique: complete subgraph of size k • k-clique community: union of all k-cliques that can be reached from each other through a series of adjacent k-cliques → they share k-1 nodes 3 -cliques and 3 -cligue percolation clusters

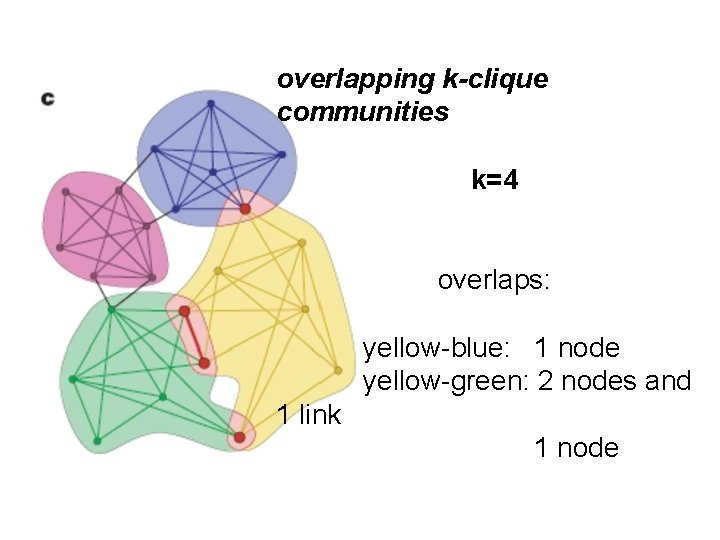
overlapping k-clique communities k=4 overlaps: yellow-blue: 1 node yellow-green: 2 nodes and 1 link 1 node

3. Finding communities Requirements: • The method of identification: – cannot be too restrictive – be based on the density of links – be local – not allowed to be any cut-node or cut-link – allow overlaps

• Algorithm: We use an exponential algorithm →it proved to be more efficient than polynomial algorithms procedure: 1. 2. Locating all cliques of the network Identifying the communities by carrying out a standard component analysis of the clique-clique overlap matrix We use the method for binary networks: undirected, unweighted links Arbitrary networks can always be transformed to binary ones: ignore any directionality keep only those links that are stronger than a treshold w*

Strategy: • according to the experience in real networks the typical size of the complete subgraphs is between 10 and 100 →( ) different k-cliques → locating the k-cliques individually and examine the adjacency between them would be extremely slow don’t look for k-cliques, rather 1. locate the large complete subgraphs 2. look for the k-clique connected subsets of given k by studying the overlap between them

Method: 1. Extract all complete subgraphs (cliques): • cliques have to be located in a decreasing order of their size (firtst of all the largest clique size have to be determined) start with this size repeatedly choose a node extract every clique of this size containing that node delete the node and its edges (will not find the same clique multiple times) when no nodes are left the clique size is decreased by one • Find the clique of size s that contains node v: construct set A A: nodes all linked to eachother initially contains v then enlarge till it reaches size s construct set B the set of nodes that are linked to each node in A but not necessarily to the nodes in B initially consists of the neighbours of v

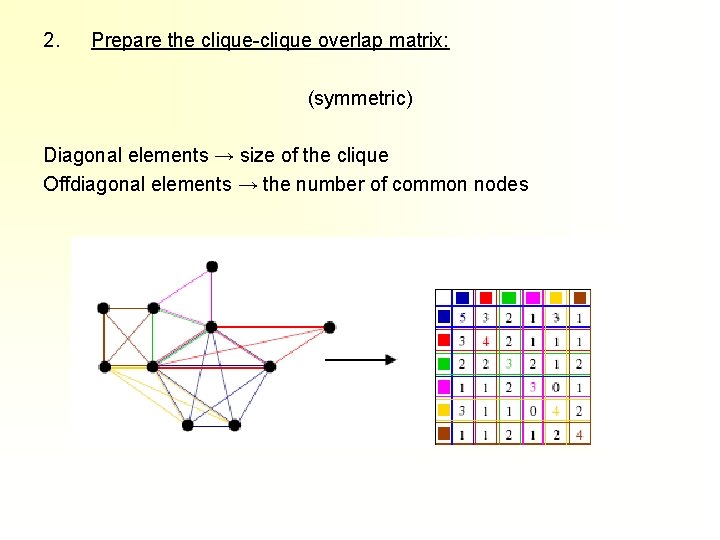
2. Prepare the clique-clique overlap matrix: (symmetric) Diagonal elements → size of the clique Offdiagonal elements → the number of common nodes

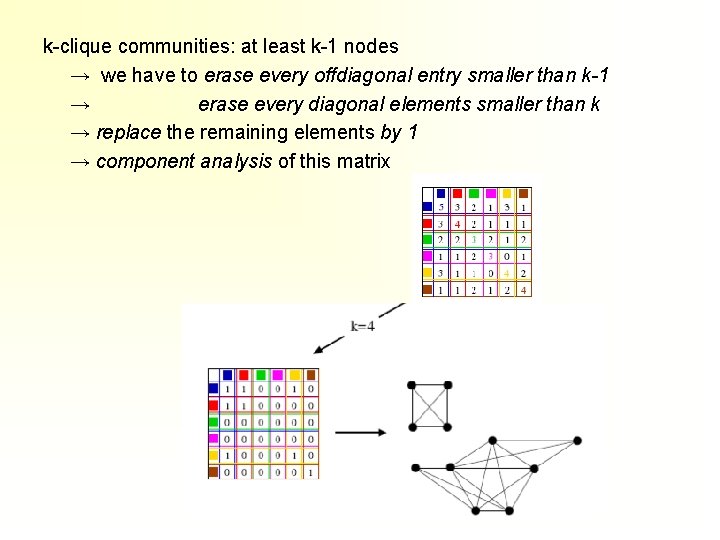
k-clique communities: at least k-1 nodes → we have to erase every offdiagonal entry smaller than k-1 → erase every diagonal elements smaller than k → replace the remaining elements by 1 → component analysis of this matrix

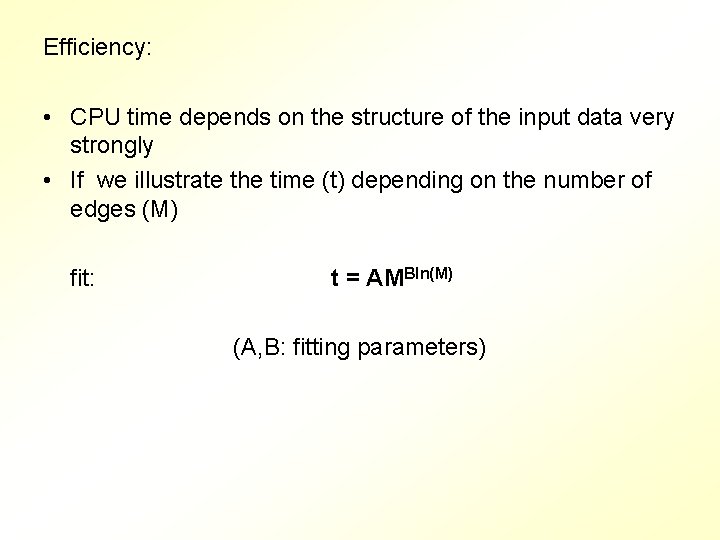
Efficiency: • CPU time depends on the structure of the input data very strongly • If we illustrate the time (t) depending on the number of edges (M) fit: t = AMBln(M) (A, B: fitting parameters)

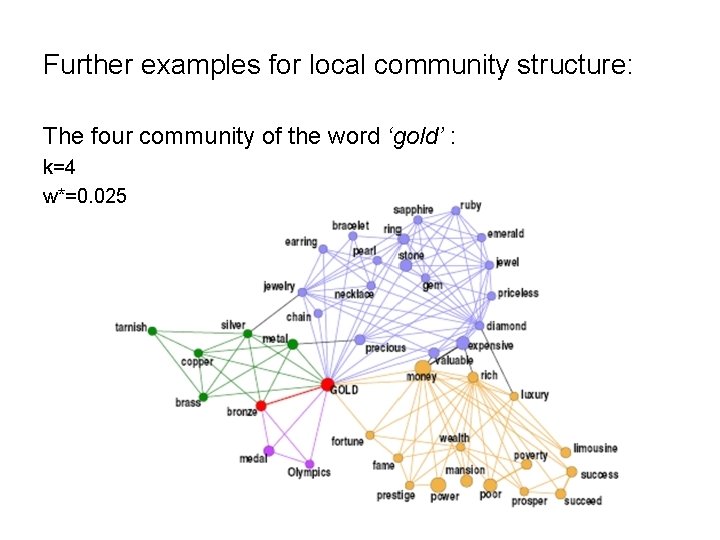
Further examples for local community structure: The four community of the word ‘gold’ : k=4 w*=0. 025

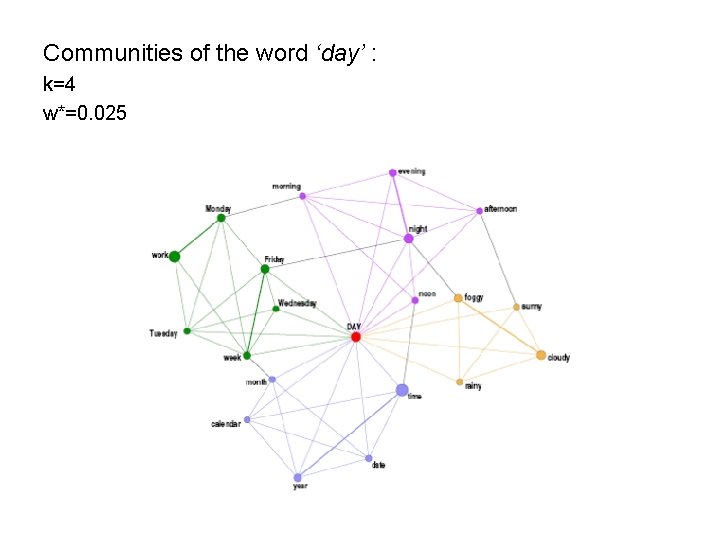
Communities of the word ‘day’ : k=4 w*=0. 025

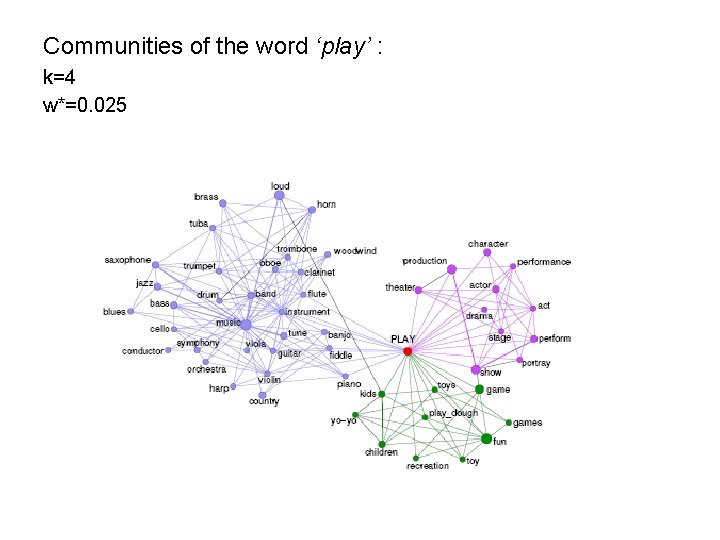
Communities of the word ‘play’ : k=4 w*=0. 025

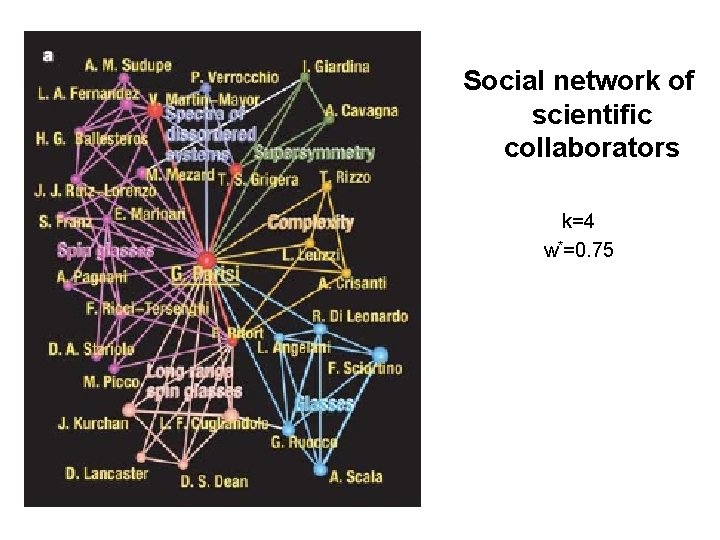
Community structure around a particular node: We should scan through some ranges of k, w* Examples: 1. Social network of scientific collaborators 2. The communities of the word ‘bright’ in the South Florida Free Association norms list 3. The communities of the protein Zds 1 in the DIP core list of the protein-protein interaction of Saccharomyces cerevisiae

Social network of scientific collaborators k=4 w*=0. 75

The communities of the word: ‘bright’ k=4 w*=0. 025

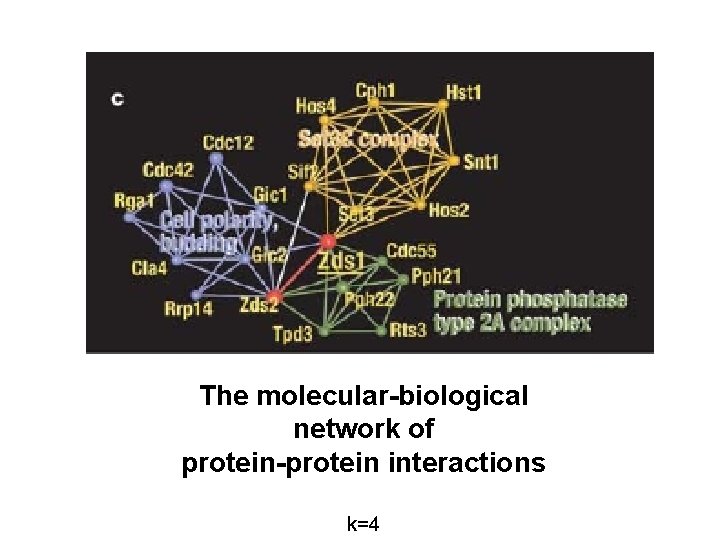
The molecular-biological network of protein-protein interactions k=4

We try to find the community of proteins based on their interaction Most proteins can be associated with • protein complexes • certain functions For some proteins no function is yet available → appearing as a part of a community can be a prediction of their functions Example: protein Ycr 072 c (essential for the viability of the cell) • there is no biological function yet available • the most important biological process for this community: ‘ribosome biogenesis/assembly’ → our protein is likely to be involved in this process

Network of the protein-protein interactions of S. cerevisiae (k=4)

Divisive and agglomerative methods Devisive methods: cut the network into smaller and smaller peaces –each node is forced to remain in only one community and becomes separated from its other communities → usually they fall apart and desappear example: ‘bright’ → stays together with the words connected to ‘light’ → most of the other communities disintegrate Agglomerative methods: • do the same in reverse direction • leads to a tree-like hierarchical rendering of the communities

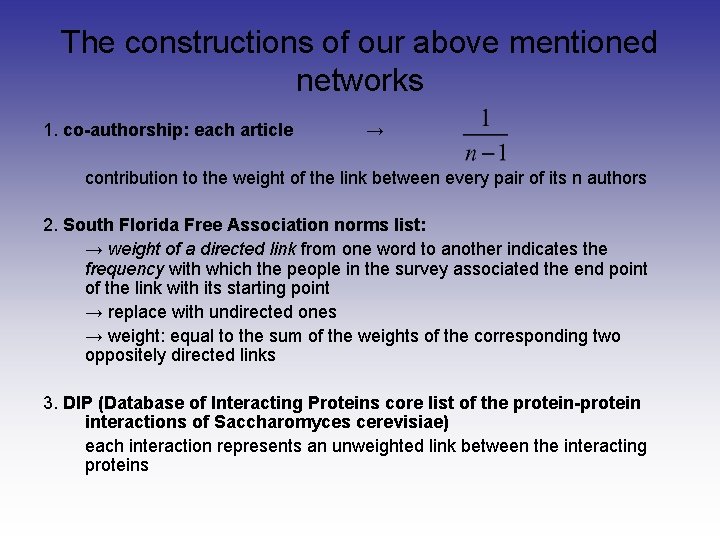
The constructions of our above mentioned networks 1. co-authorship: each article → contribution to the weight of the link between every pair of its n authors 2. South Florida Free Association norms list: → weight of a directed link from one word to another indicates the frequency with which the people in the survey associated the end point of the link with its starting point → replace with undirected ones → weight: equal to the sum of the weights of the corresponding two oppositely directed links 3. DIP (Database of Interacting Proteins core list of the protein-protein interactions of Saccharomyces cerevisiae) each interaction represents an unweighted link between the interacting proteins

4. Statistical features Values of k, w*: Purpose: we would like to analyse the statistical properties of the community structure of the entire network → finding a community structure that is as highly structured as possible it leads us to the percolation phenomenon: If the number of links is increased above a critical point a giant component appears.

Approach critical point! • for each value of k (typ. 3 -6) we lower the treshold w* until the largest community becomes twice as big as the second largest one find as many communities as possible, but – no giant community that smears out the details of the community structure by merging many smaller communities • f*: the fraction of links stronger than w* – use those k values for which f* is not too small (smaller than 0. 5) • • • co-authorship: protein interaction network: word-association: k=6 k=5 k=4 f*= 0. 93 f*= 0. 75 f*= 0. 67

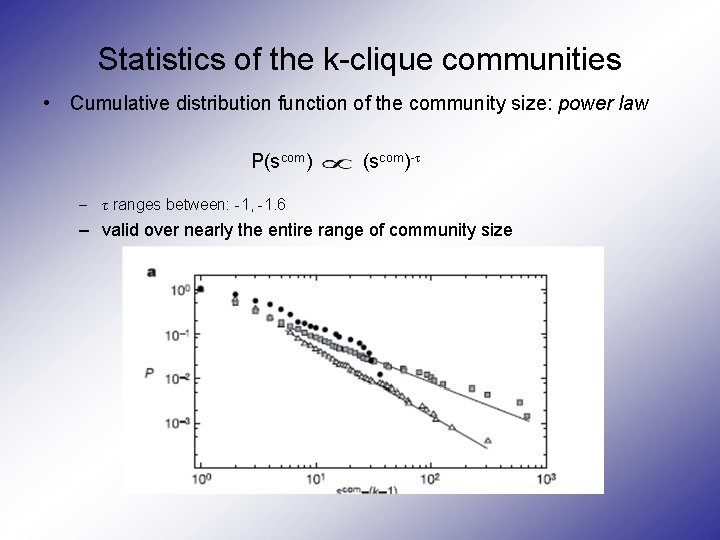
Statistics of the k-clique communities • Cumulative distribution function of the community size: power law P(scom)-τ – τ ranges between: -1, -1. 6 – valid over nearly the entire range of community size

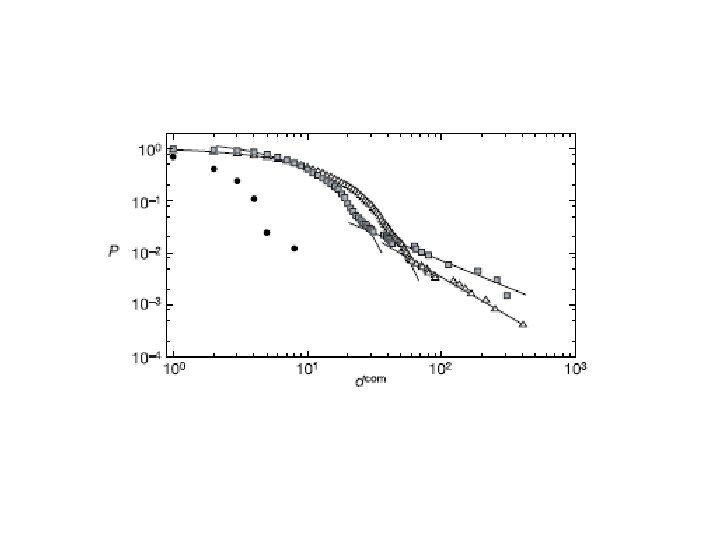
• The cumulative distribution of the community degree: – starts exponentionally then crosses over to a power law • exponentional decay: P(dcom) most of the communities have a size of the order of k and their distribution dominates this part of the curve → a characteristic scale appears d 0 com ≈ kδ • power-law tail: P(dcom) –τ on average each node of a community has a contribution of δ to the community degree → this power law tail is proportional to that of the community size distribution


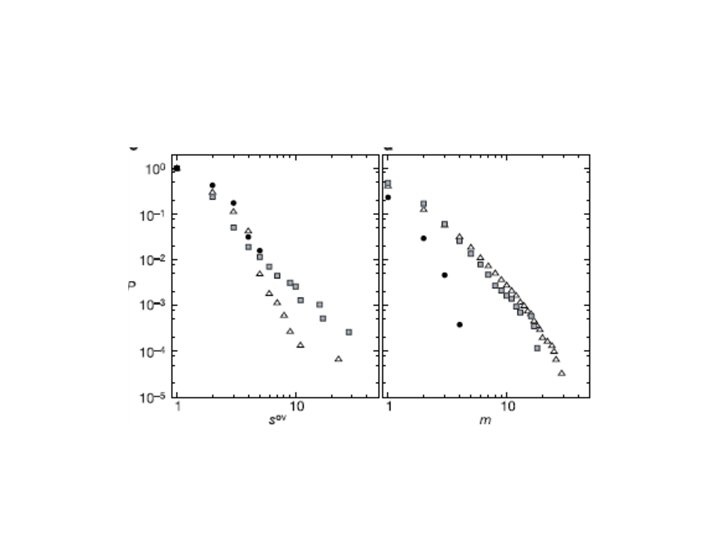
• The cumulative distribution of the overlap size: – close to a power law – large exponent – there is no characteristic overlap size in the network • The cumulative distribution of the membership number: P(m) – a node can belong to several communities – collaboration, word-association: • no characteristic value • the data are close to a power-law dependence, large exponent – protein-protein interaction: the largest membership number is only 4 (consistent with the also short distribution of its community degree)


From statistical features: • two communities overlapping with a given community are likely to overlap with each other as well ( average clustering coefficient is high ) • Specific scaling of P(dcom): the signature of the hierarchical nature of the system (the network of the communities still exhibits a degreedistribution with a fat tail, a characteristic scale appears below which the distribution is exponential) Complex systems have different levels of organization with units specific to each level

5. The importance of observing networks • Community structure → prediction of some essential features of the system possibility to ‘zoom’ in on a unit and uncover its communities → interpret the local organization of large networks → predict how the modular structure changes if a unit is removed We can simultaneously look at the network at a higher level of organization and locate the communities.