The Organic Chemistry of EnzymeCatalyzed Reactions Chapter 2








![Evidence for Histidine Participation substrate inactivator (TPCK) With [14 C]TPCK get 1 equiv. [14 Evidence for Histidine Participation substrate inactivator (TPCK) With [14 C]TPCK get 1 equiv. [14](https://slidetodoc.com/presentation_image_h2/ceaa89d8c1a2586db71b194813c504d7/image-18.jpg)



















- Slides: 77

The Organic Chemistry of Enzyme-Catalyzed Reactions Chapter 2 Group Transfer Reactions: Hydrolysis, Amination, Phosphorylation

Hydrolysis Reactions Amide Hydrolysis Peptidases (proteases if protein hydrolysis involved) catalyze the hydrolysis of peptide bonds

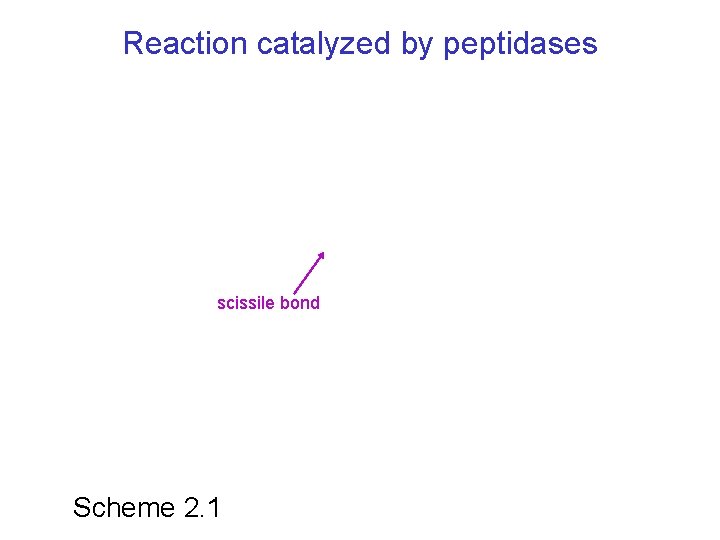
Reaction catalyzed by peptidases scissile bond Scheme 2. 1

Classifications of peptidases Figure 2. 1

Endopeptidases • Representative example is -chymotrypsin • Regiospecifically hydrolyzes peptide bonds of the aromatic acids • P 1 -chymotrypsin is Phe, Tyr, and Trp • P 1 for trypsin is Arg and Lys

Endopeptidase Mechanism for -chymotrypsin showing catalytic triad Scheme 2. 2

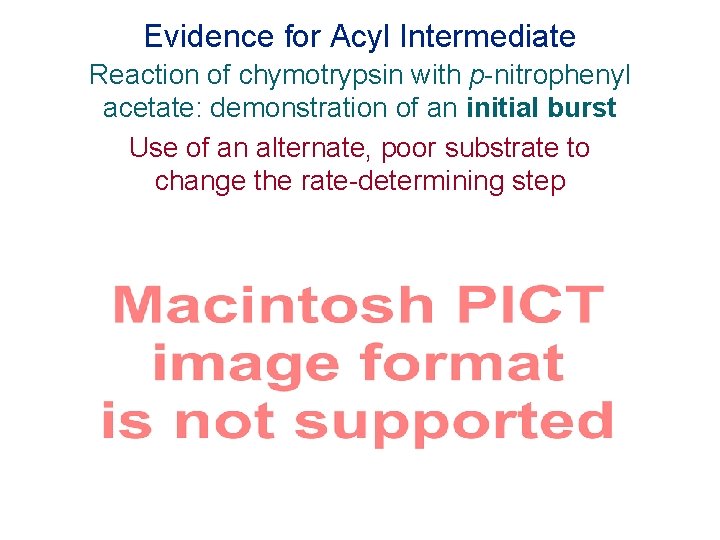
Evidence for Acyl Intermediate Reaction of chymotrypsin with p-nitrophenyl acetate: demonstration of an initial burst Use of an alternate, poor substrate to change the rate-determining step Figure 2. 2

Typical enzyme reaction in which the first step is fast Scheme 2. 3

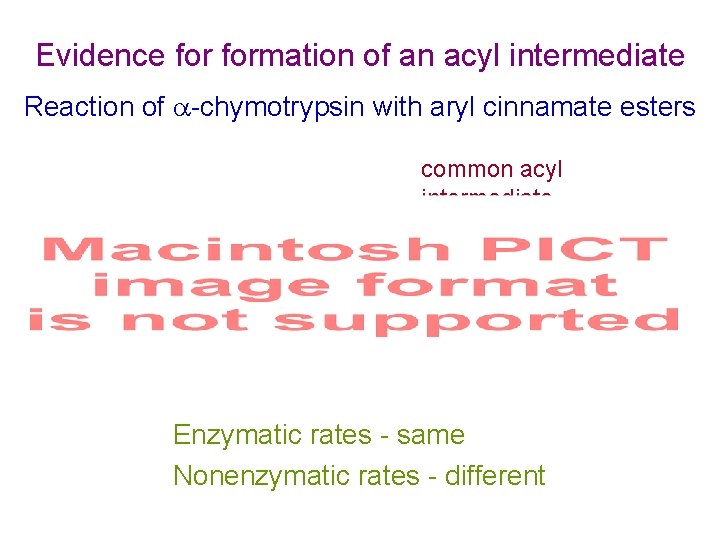
Evidence formation of an acyl intermediate Reaction of -chymotrypsin with aryl cinnamate esters common acyl intermediate Scheme 2. 4 Enzymatic rates - same Nonenzymatic rates - different

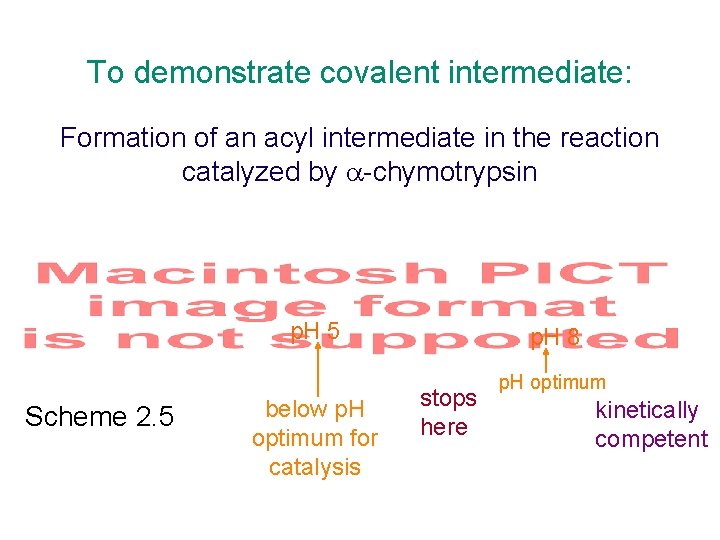
To demonstrate covalent intermediate: Formation of an acyl intermediate in the reaction catalyzed by -chymotrypsin p. H 5 Scheme 2. 5 below p. H optimum for catalysis p. H 8 stops here p. H optimum kinetically competent

Gel Filtration (aromatic amino acids in enzyme) excess substrate Figure 2. 3

Reactivation of acetylchymotrypsin by hydroxylamine To support formation of acetylchymotrypsin reactivated enzyme Isolate and characterize Scheme 2. 6

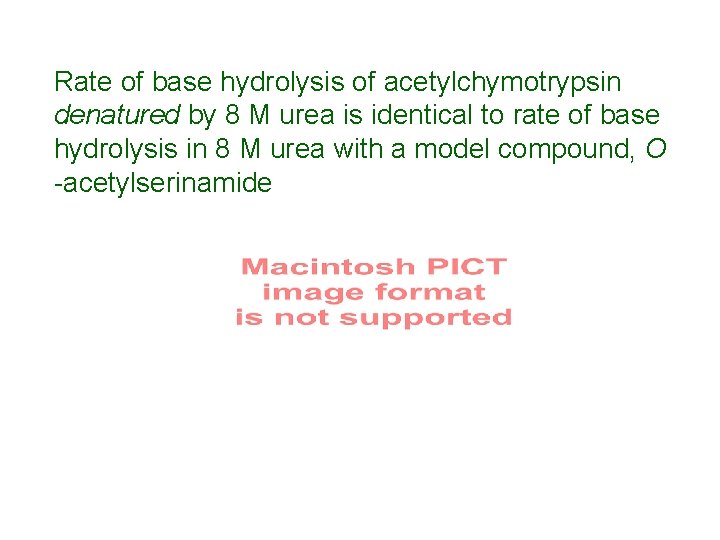
Rate of base hydrolysis of acetylchymotrypsin denatured by 8 M urea is identical to rate of base hydrolysis in 8 M urea with a model compound, O -acetylserinamide

Reaction of -chymotrypsin with an organophosphofluoridate affinity labeling agent To show involvement of a serine residue at the active site affinity labeling agent Scheme 2. 7

Affinity labeling agent Kinetics of affinity labeling of enzymes substrate protection Scheme 2. 8

• Irreversible inhibitors exhibit time-dependent inhibition Reaction after E • I complex formation is rate limiting; therefore, time dependent

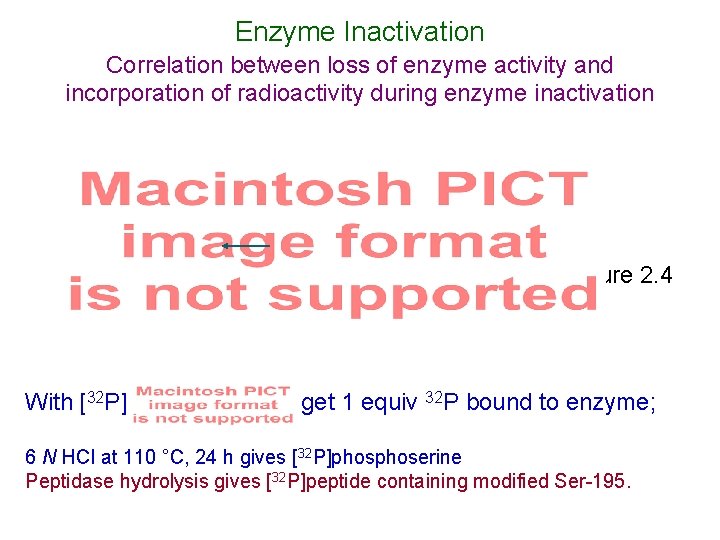
Enzyme Inactivation Correlation between loss of enzyme activity and incorporation of radioactivity during enzyme inactivation loss of enzyme activity and incorporation of radioactivity correspond (1 : 1 inactivator : enzyme) With [32 P] Figure 2. 4 get 1 equiv 32 P bound to enzyme; 6 N HCl at 110 °C, 24 h gives [32 P]phoserine Peptidase hydrolysis gives [32 P]peptide containing modified Ser-195.
![Evidence for Histidine Participation substrate inactivator TPCK With 14 CTPCK get 1 equiv 14 Evidence for Histidine Participation substrate inactivator (TPCK) With [14 C]TPCK get 1 equiv. [14](https://slidetodoc.com/presentation_image_h2/ceaa89d8c1a2586db71b194813c504d7/image-18.jpg)
Evidence for Histidine Participation substrate inactivator (TPCK) With [14 C]TPCK get 1 equiv. [14 C] bound; pepsin hydrolysis gives a [14 C] peptide with His-57 modified

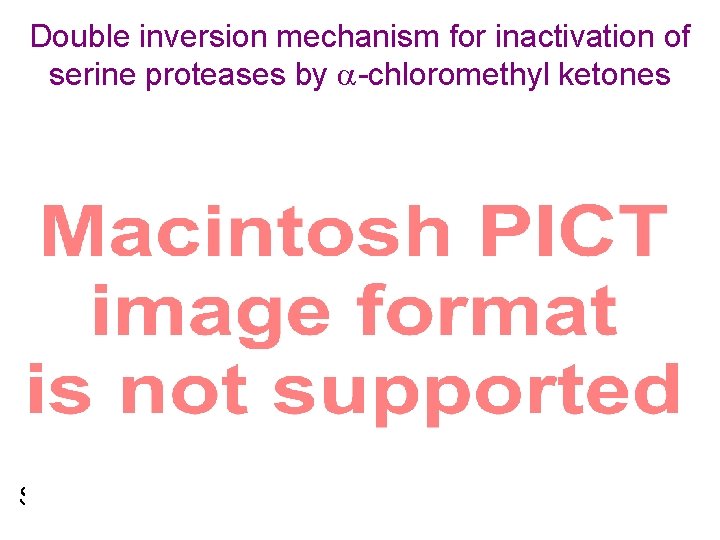
Mechanism of inactivation of chymotrypsin by -chloromethyl ketones -chymotrypsin (side reaction) (S)-N-Ac-L-Ala-L-Phe No hydrolysis product in absence of enzyme (nonenzyme control) Evidence against a single SN 2 reaction Same stereochemistry as 2. 13

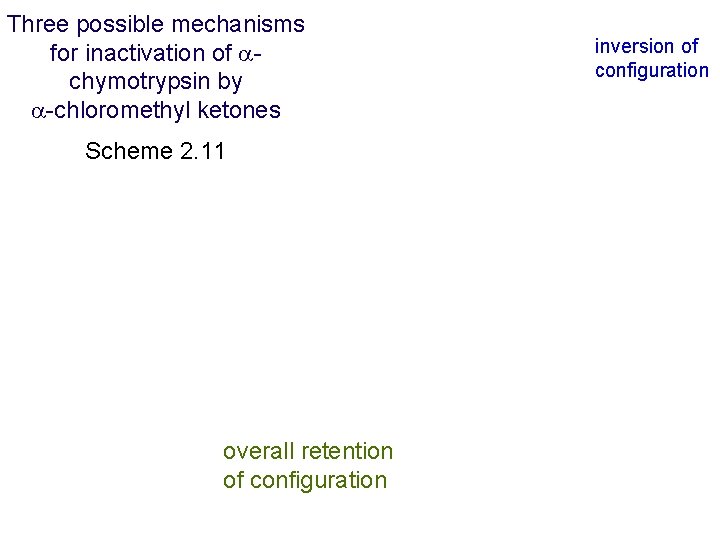
Double inversion mechanism for inactivation of serine proteases by -chloromethyl ketones Scheme 2. 10

Three possible mechanisms for inactivation of chymotrypsin by -chloromethyl ketones Scheme 2. 11 overall retention of configuration inversion of configuration

-Chymotrypsin was inactivated by 2. 20, and X-ray crystal structure showed His 57 alkylated with stereochemistry retained

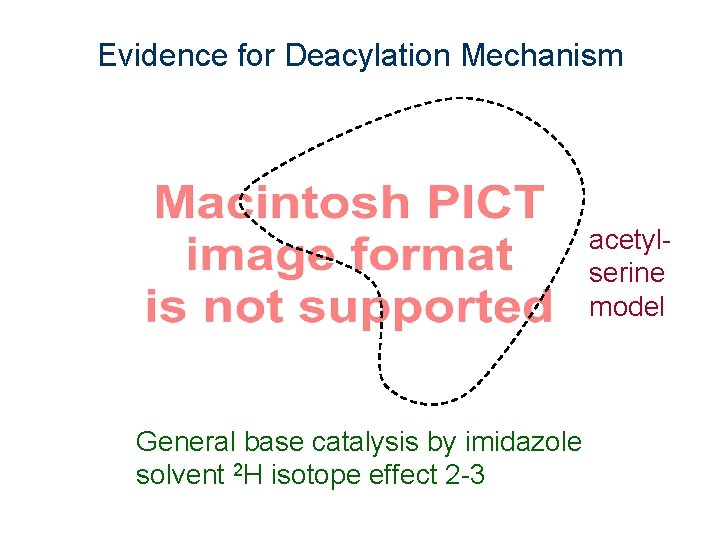
Evidence for Deacylation Mechanism acetylserine model General base catalysis by imidazole solvent 2 H isotope effect 2 -3

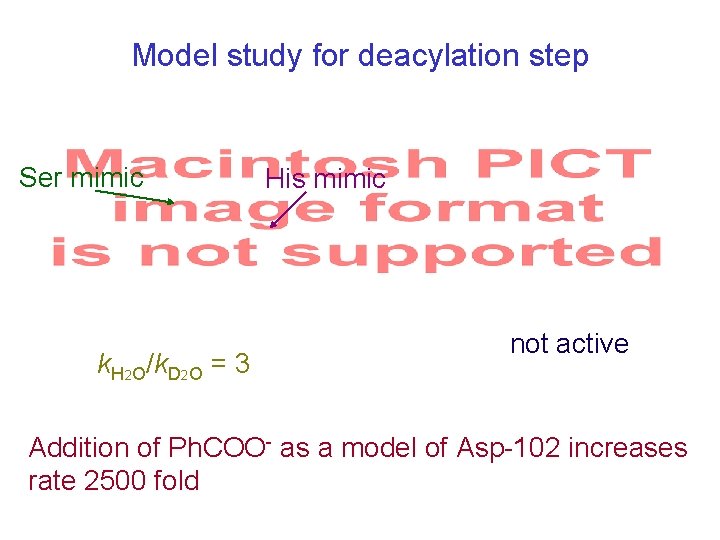
Model study for deacylation step Ser mimic k. H 2 O/k. D 2 O = 3 His mimic not active Addition of Ph. COO- as a model of Asp-102 increases rate 2500 fold

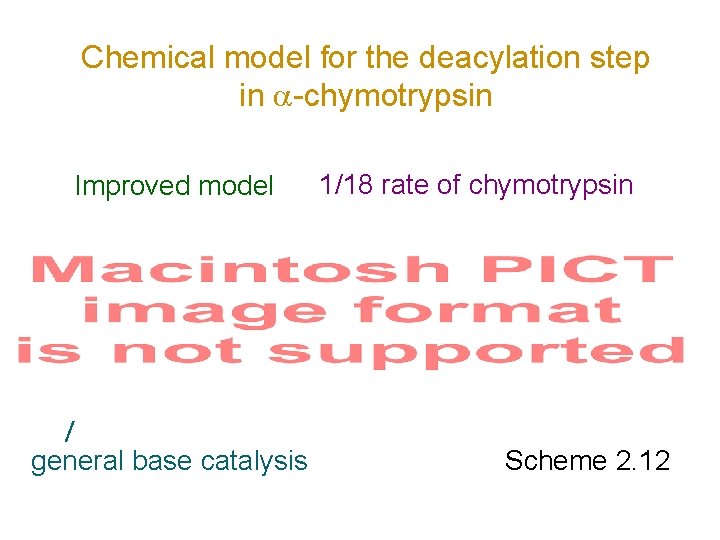
Chemical model for the deacylation step in -chymotrypsin Improved model general base catalysis 1/18 rate of chymotrypsin Scheme 2. 12


Aspartate Protease Proposed mechanism for HIV-1 protease Note: General acidbase catalysis, not covalent catalysis Scheme 2. 14

Carboxypeptidases (an exopeptidase) Affinity labeling agent for CPA labels Glu-270

General base catalytic mechanism for carboxypeptidase A Scheme 2. 15 Zn++ is a cofactor

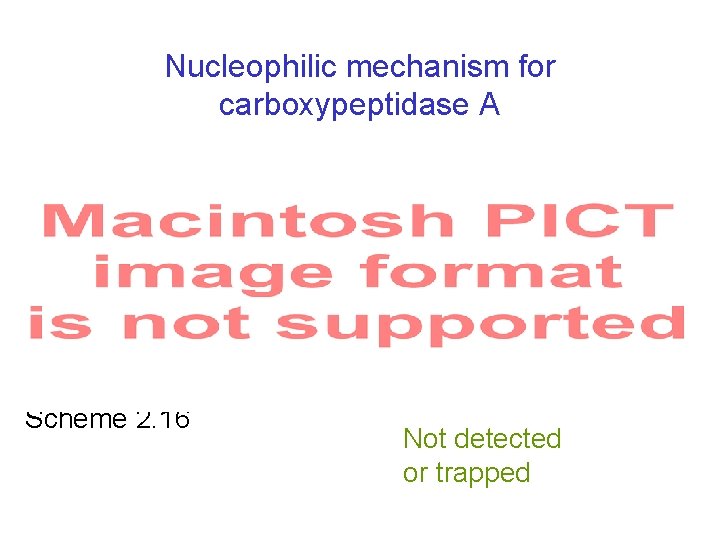
Nucleophilic mechanism for carboxypeptidase A Scheme 2. 16 Not detected or trapped

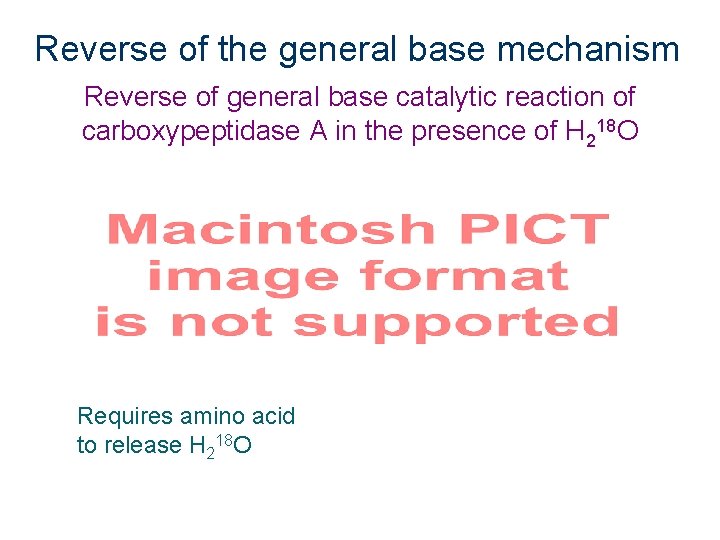
Principle of Microscopic Reversibility For any reversible reaction, the mechanism in the reverse direction must be identical to that in the forward reaction (only reversed) This can be a valuable approach to study enzyme mechanisms.

Reverse of the general base mechanism Reverse of general base catalytic reaction of carboxypeptidase A in the presence of H 218 O Scheme 2. 17 Requires amino acid to release H 218 O

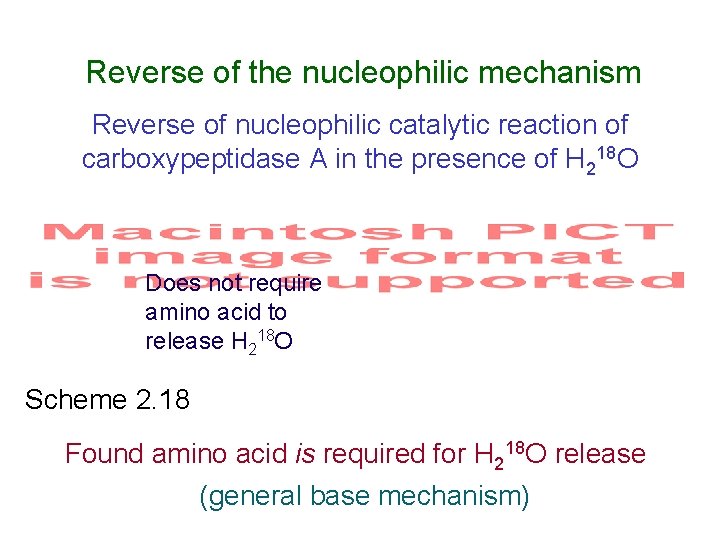
Reverse of the nucleophilic mechanism Reverse of nucleophilic catalytic reaction of carboxypeptidase A in the presence of H 218 O Does not require amino acid to release H 218 O Scheme 2. 18 Found amino acid is required for H 218 O release (general base mechanism)

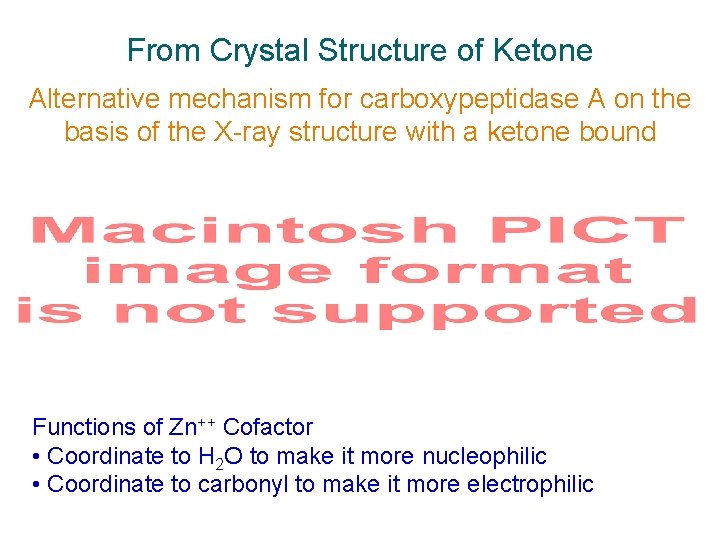
From Crystal Structure of Ketone Alternative mechanism for carboxypeptidase A on the basis of the X-ray structure with a ketone bound Scheme 2. 19 Functions of Zn++ Cofactor • Coordinate to H 2 O to make it more nucleophilic • Coordinate to carbonyl to make it more electrophilic

Typical esterase mechanism Scheme 2. 20 Covalent catalytic mechanism

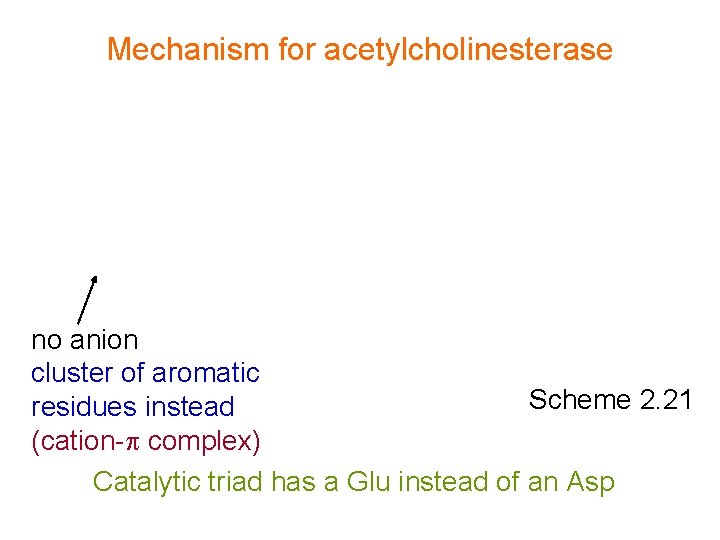
Mechanism for acetylcholinesterase no anion cluster of aromatic Scheme 2. 21 residues instead (cation- complex) Catalytic triad has a Glu instead of an Asp

Favored enantiomer substrate for lipases

An example of the enantioselectivity of lipases/esterases Scheme 2. 22 Useful for chiral resolutions of alcohols

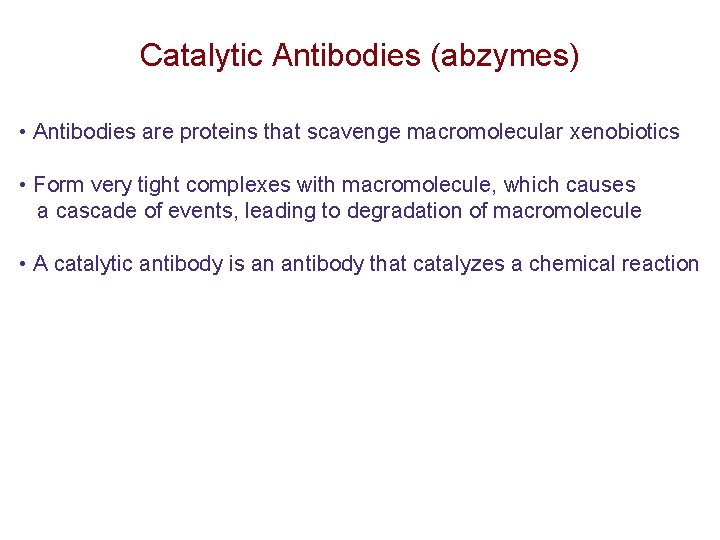
Catalytic Antibodies (abzymes) • Antibodies are proteins that scavenge macromolecular xenobiotics • Form very tight complexes with macromolecule, which causes a cascade of events, leading to degradation of macromolecule • A catalytic antibody is an antibody that catalyzes a chemical reaction

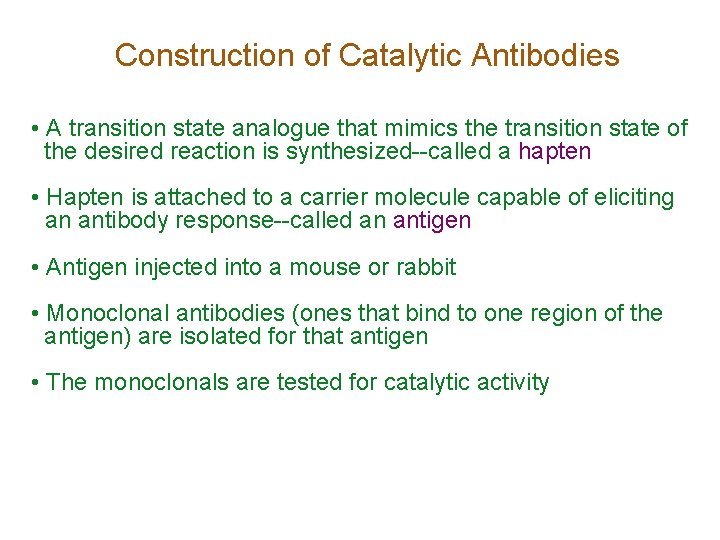
Construction of Catalytic Antibodies • A transition state analogue that mimics the transition state of the desired reaction is synthesized--called a hapten • Hapten is attached to a carrier molecule capable of eliciting an antibody response--called an antigen • Antigen injected into a mouse or rabbit • Monoclonal antibodies (ones that bind to one region of the antigen) are isolated for that antigen • The monoclonals are tested for catalytic activity

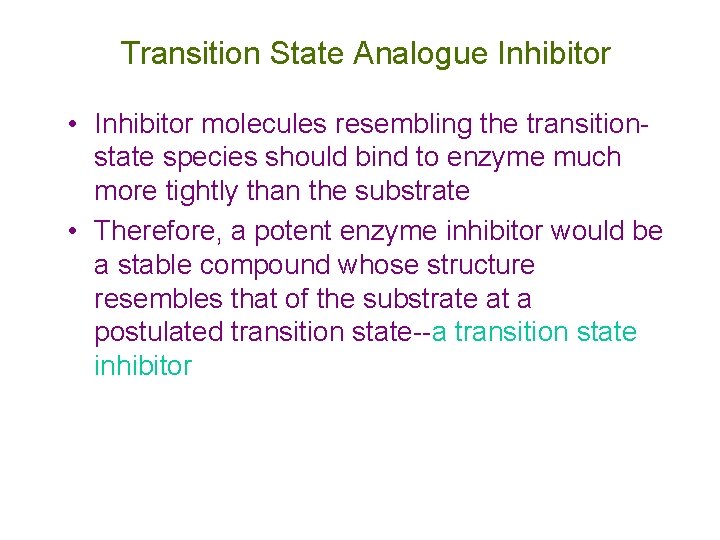
Transition State Analogue Inhibitor • Inhibitor molecules resembling the transitionstate species should bind to enzyme much more tightly than the substrate • Therefore, a potent enzyme inhibitor would be a stable compound whose structure resembles that of the substrate at a postulated transition state--a transition state inhibitor

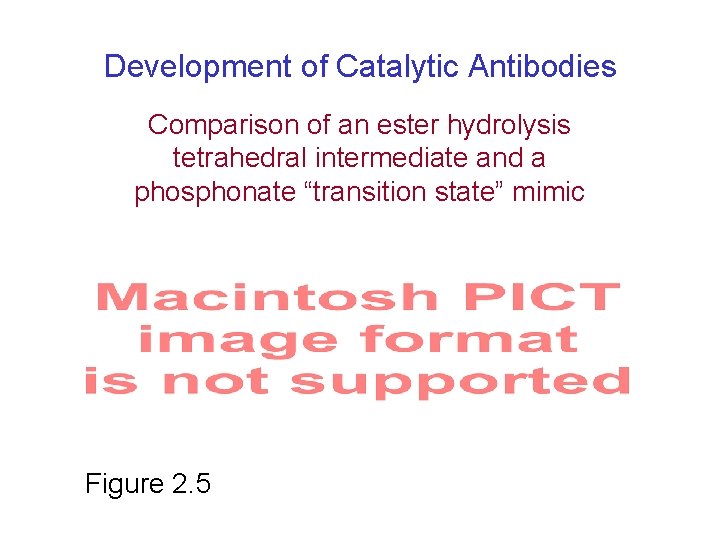
Development of Catalytic Antibodies Comparison of an ester hydrolysis tetrahedral intermediate and a phosphonate “transition state” mimic Figure 2. 5

mimics tetrahedral intermediate in ester hydrolysis X = OH hapten X = macromolecule antigen (elicits antibody response)

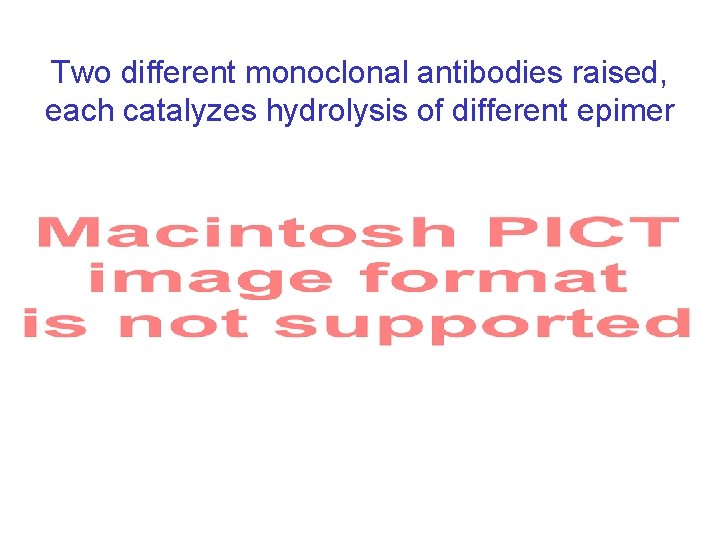
Two different monoclonal antibodies raised, each catalyzes hydrolysis of different epimer R 1 = Bn R 1 = H R 2 = Bn

Aminations

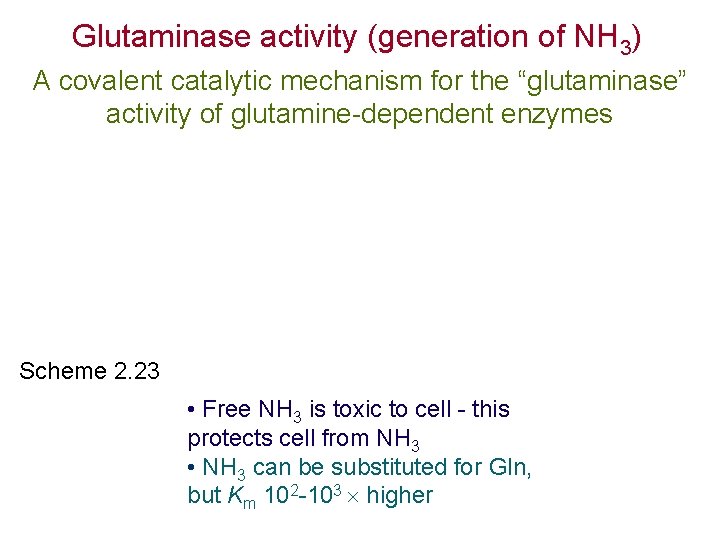
Glutaminase activity (generation of NH 3) A covalent catalytic mechanism for the “glutaminase” activity of glutamine-dependent enzymes Scheme 2. 23 • Free NH 3 is toxic to cell - this protects cell from NH 3 • NH 3 can be substituted for Gln, but Km 102 -103 higher

Evidence for covalent catalysis Evidence for -glutamyl enzyme intermediate in glutamine-dependent enzyme Scheme 2. 24

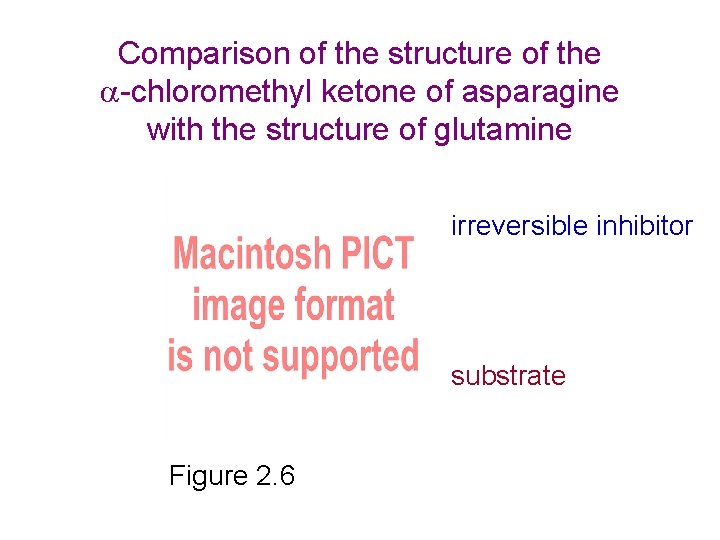
Comparison of the structure of the -chloromethyl ketone of asparagine with the structure of glutamine irreversible inhibitor substrate Figure 2. 6

modify Cys residue Blocks enzyme reaction with Gln, but not with NH 3; therefore 2 binding sites

Mechanism-based inactivators of Gln-dependent enzymes Mechanism-based inactivator • Unreactive compound whose structure resembles the substrate (or product) for an enzyme • Acts like a substrate and is converted into a species that inactivates the enzyme • Cannot escape enzyme until it inactivates it

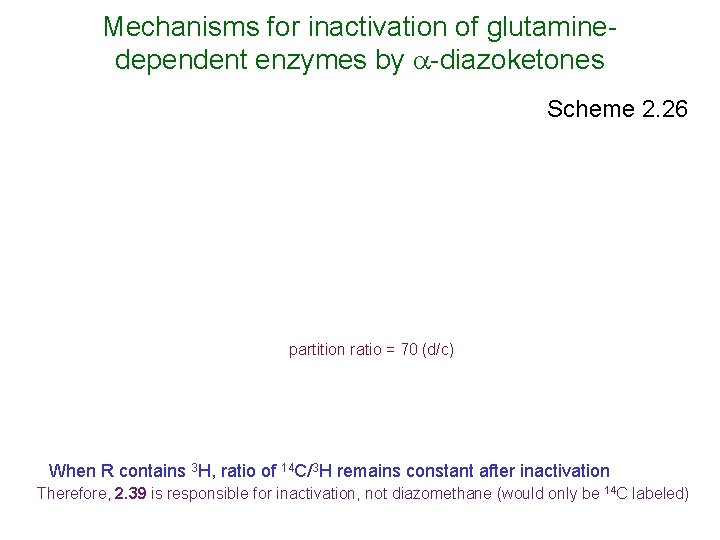
Mechanisms for inactivation of glutaminedependent enzymes by -diazoketones Scheme 2. 26 partition ratio = 70 (d/c) When R contains 3 H, ratio of 14 C/3 H remains constant after inactivation Therefore, 2. 39 is responsible for inactivation, not diazomethane (would only be 14 C labeled)

Kinetics for mechanism-based inactivation Scheme 2. 25 partition ratio = k 3/k 4 Ideally would be 0

Acceptor reactions are mostly ATP-dependent An example where no ATP is required Amination reaction catalyzed by glutamine phosphoribosyldiphosphate amidotransferase good leaving group Scheme 2. 27 SN 2 -like reaction 5 -phosphoribosyl-1 -diphosphate amidotransferase

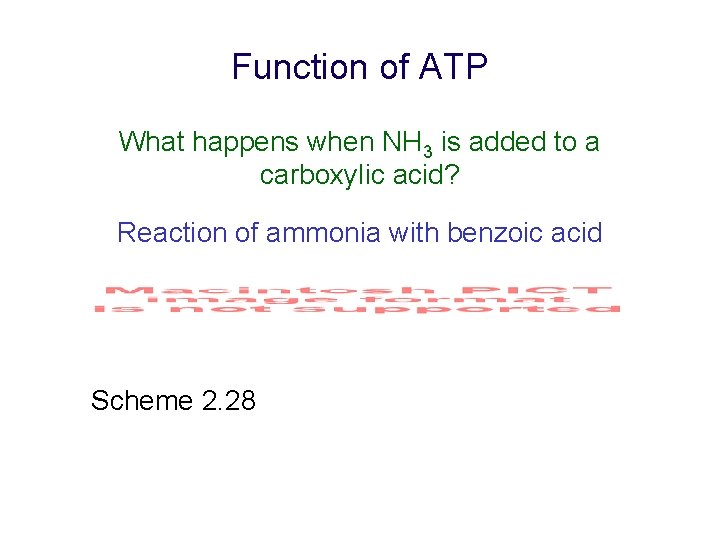
Function of ATP What happens when NH 3 is added to a carboxylic acid? Reaction of ammonia with benzoic acid Scheme 2. 28

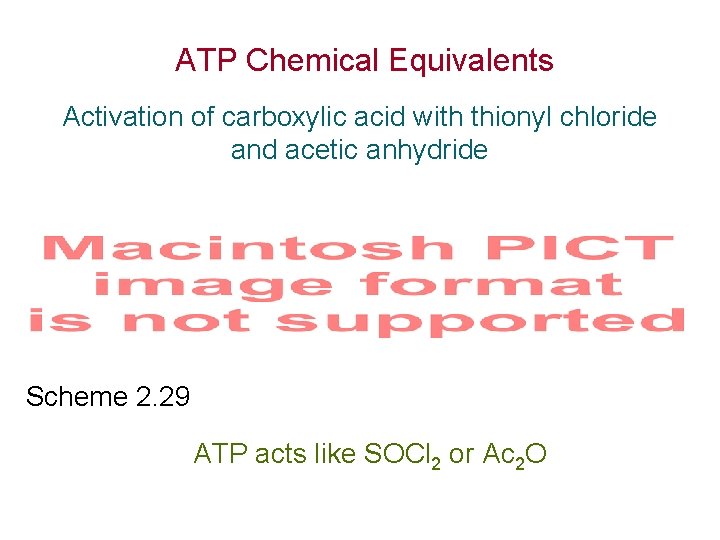
ATP Chemical Equivalents Activation of carboxylic acid with thionyl chloride and acetic anhydride Scheme 2. 29 ATP acts like SOCl 2 or Ac 2 O

Electrophilic sites on ATP Figure 2. 7 Requires Mg 2+ for activity (coordinates to phosphate oxyanions)

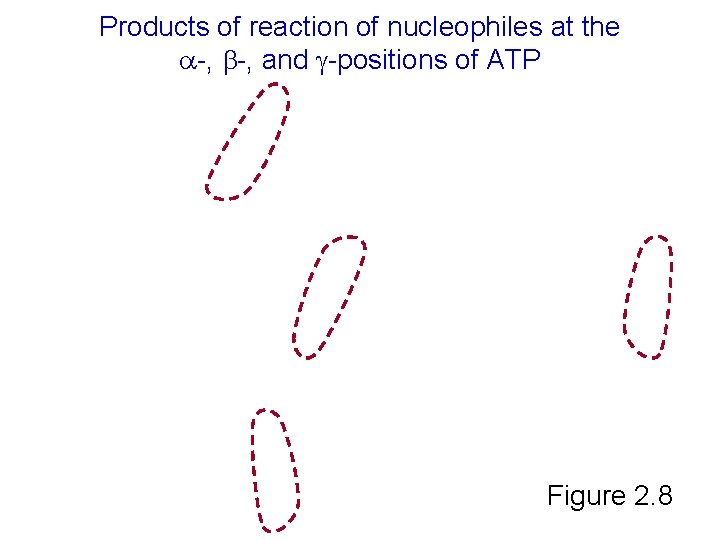
Products of reaction of nucleophiles at the -, and -positions of ATP Figure 2. 8

Reaction Catalyzed by Asparagine Synthetase Scheme 2. 30

Two possible modes of attack to give AMP + PPi Activation of aspartate by ATP followed by reaction with ammonia generated from glutamine Scheme 2. 31

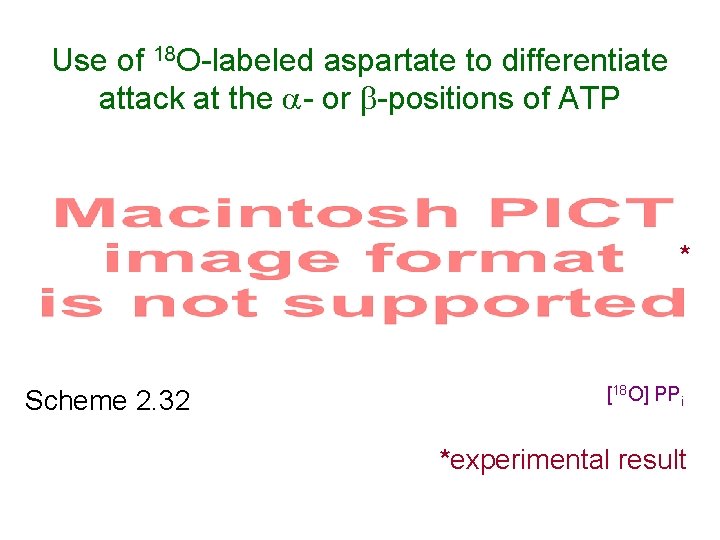
Use of 18 O-labeled aspartate to differentiate attack at the - or -positions of ATP [18 O] AMP* Scheme 2. 32 [18 O] PPi *experimental result

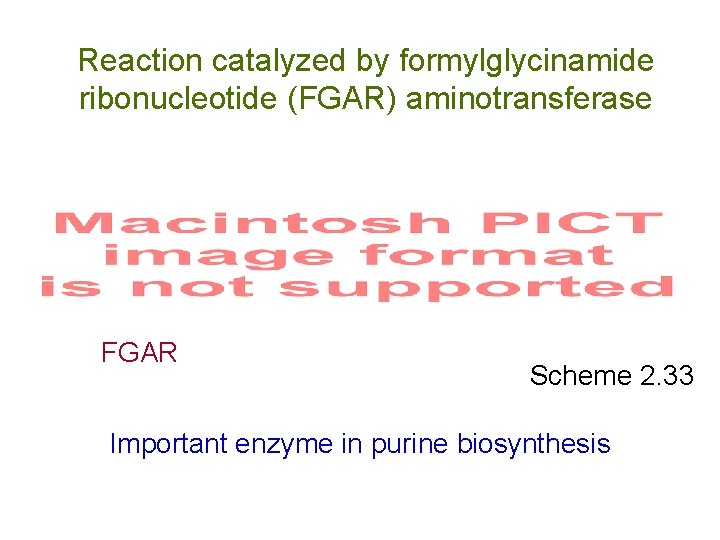
Reaction catalyzed by formylglycinamide ribonucleotide (FGAR) aminotransferase FGAR Scheme 2. 33 Important enzyme in purine biosynthesis

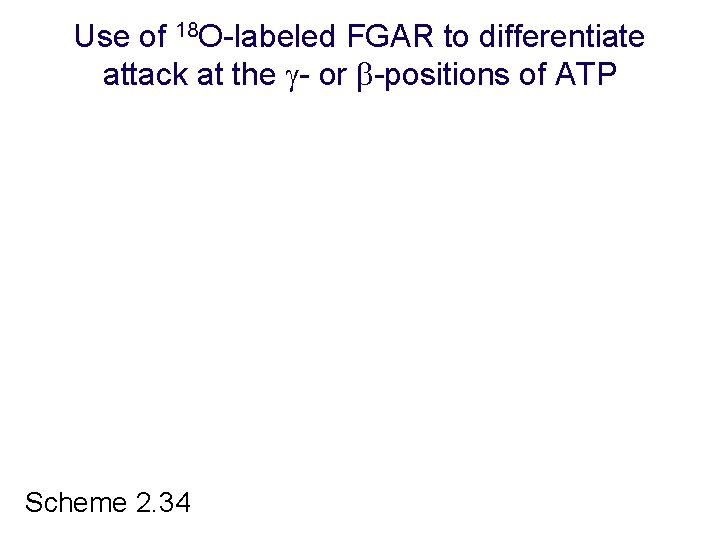
Use of 18 O-labeled FGAR to differentiate attack at the - or -positions of ATP Scheme 2. 34

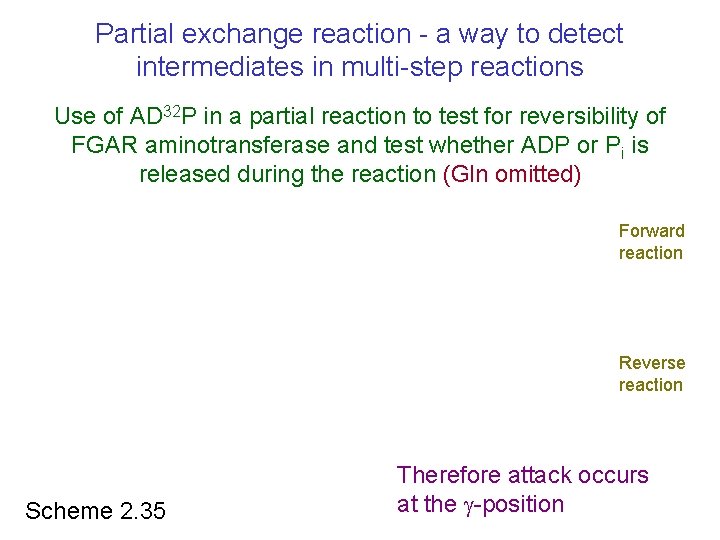
Partial exchange reaction - a way to detect intermediates in multi-step reactions Use of AD 32 P in a partial reaction to test for reversibility of FGAR aminotransferase and test whether ADP or Pi is released during the reaction (Gln omitted) Forward reaction Reverse reaction Scheme 2. 35 Therefore attack occurs at the -position

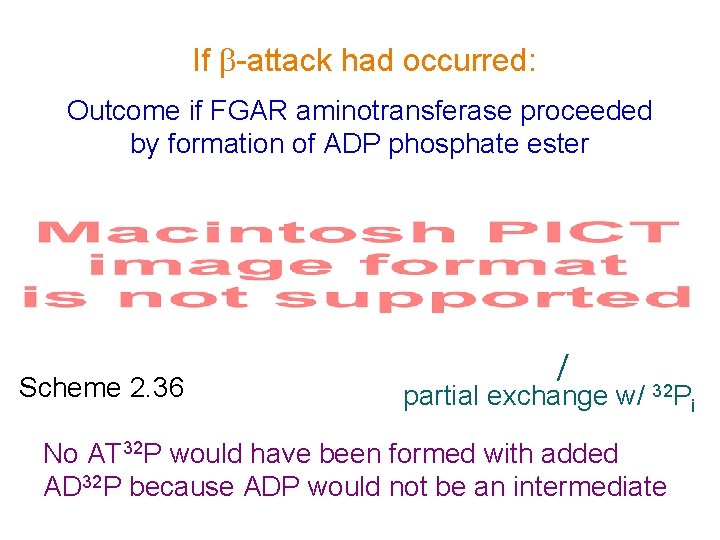
If -attack had occurred: Outcome if FGAR aminotransferase proceeded by formation of ADP phosphate ester Scheme 2. 36 partial exchange w/ 32 Pi No AT 32 P would have been formed with added AD 32 P because ADP would not be an intermediate

If neither experiment leads to incorporation of 32 P into the ATP, it does not mean that neither intermediate is formed • Assumed enzyme followed an ordered mechanism and that the first partial reaction could proceed in the absence of glutamine: Maybe enzyme needs the glutamine to be bound before activation occurs Binding of glutamine may cause a conformational change that sets up binding site for FGAR and ATP • Another potential problem - ADP generated in the first partial reaction may bind very tightly, so dissociation and exchange with AD 32 P do not occur

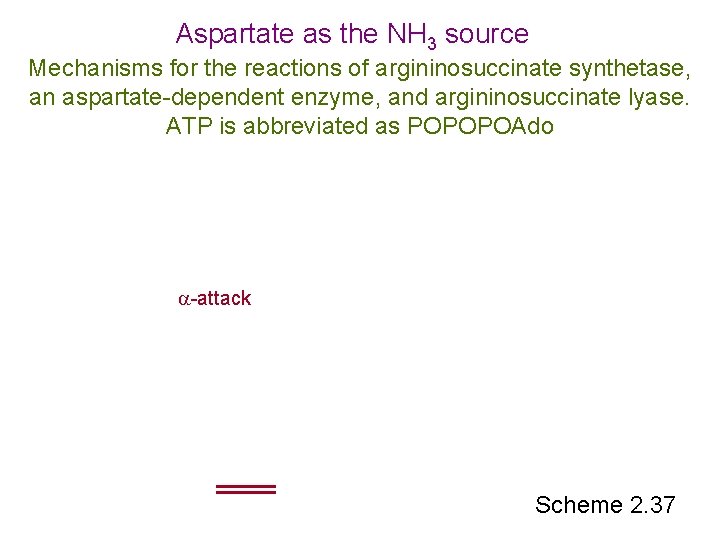
Aspartate as the NH 3 source Mechanisms for the reactions of argininosuccinate synthetase, an aspartate-dependent enzyme, and argininosuccinate lyase. ATP is abbreviated as POPOPOAdo -attack Scheme 2. 37

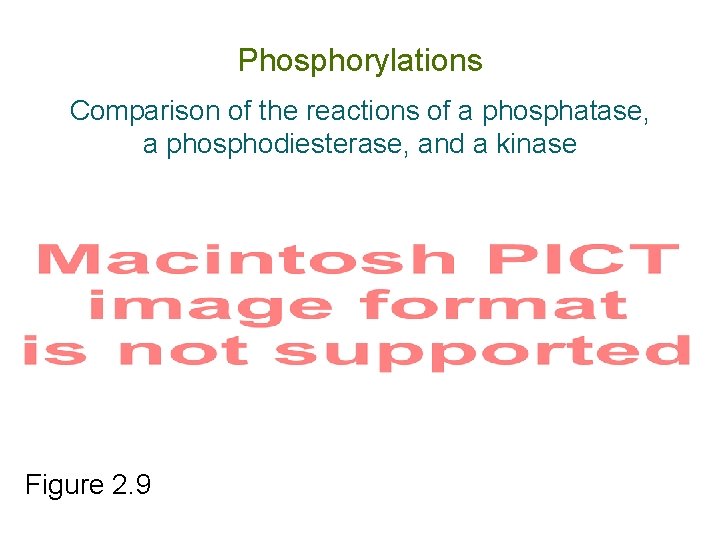
Phosphorylations Comparison of the reactions of a phosphatase, a phosphodiesterase, and a kinase Figure 2. 9

Three general mechanisms for phosphatases metaphosphate Scheme 2. 38

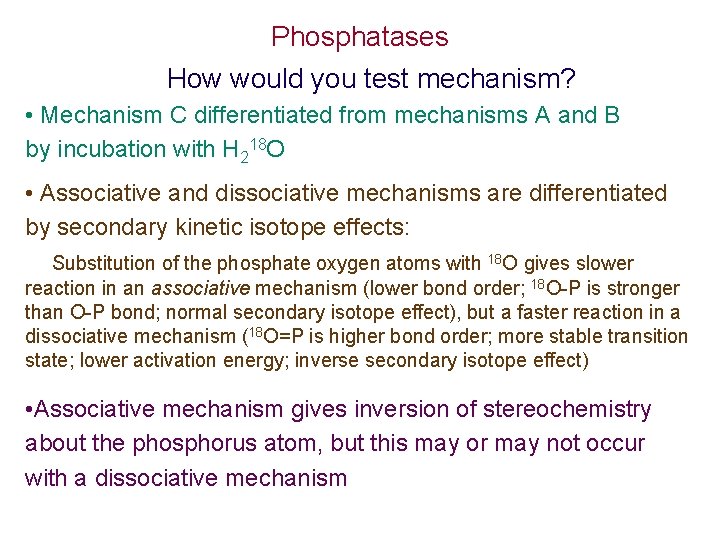
Phosphatases How would you test mechanism? • Mechanism C differentiated from mechanisms A and B by incubation with H 218 O • Associative and dissociative mechanisms are differentiated by secondary kinetic isotope effects: Substitution of the phosphate oxygen atoms with 18 O gives slower reaction in an associative mechanism (lower bond order; 18 O-P is stronger than O-P bond; normal secondary isotope effect), but a faster reaction in a dissociative mechanism (18 O=P is higher bond order; more stable transition state; lower activation energy; inverse secondary isotope effect) • Associative mechanism gives inversion of stereochemistry about the phosphorus atom, but this may or may not occur with a dissociative mechanism

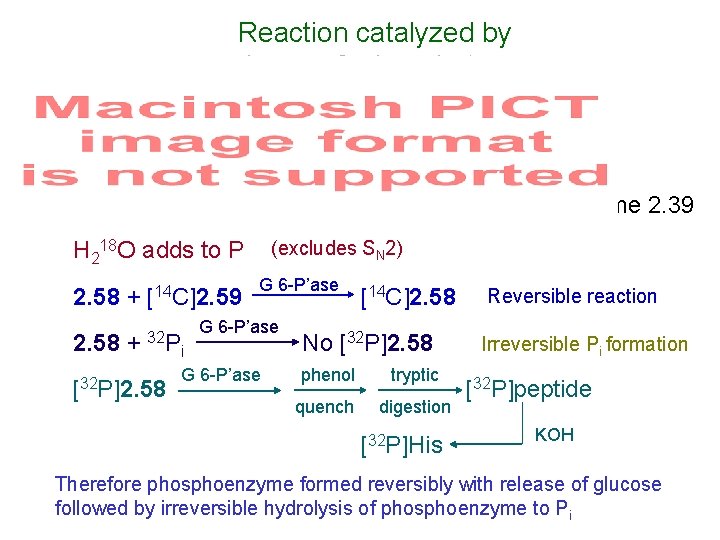
Reaction catalyzed by glucose 6 -phosphatase Scheme 2. 39 (excludes SN 2) H 218 O adds to P 2. 58 + [14 C]2. 59 2. 58 + 32 P [32 P]2. 58 G 6 -P’ase [14 C]2. 58 i No [32 P]2. 58 G 6 -P’ase phenol tryptic quench digestion [32 P]His Reversible reaction Irreversible Pi formation [32 P]peptide KOH Therefore phosphoenzyme formed reversibly with release of glucose followed by irreversible hydrolysis of phosphoenzyme to Pi

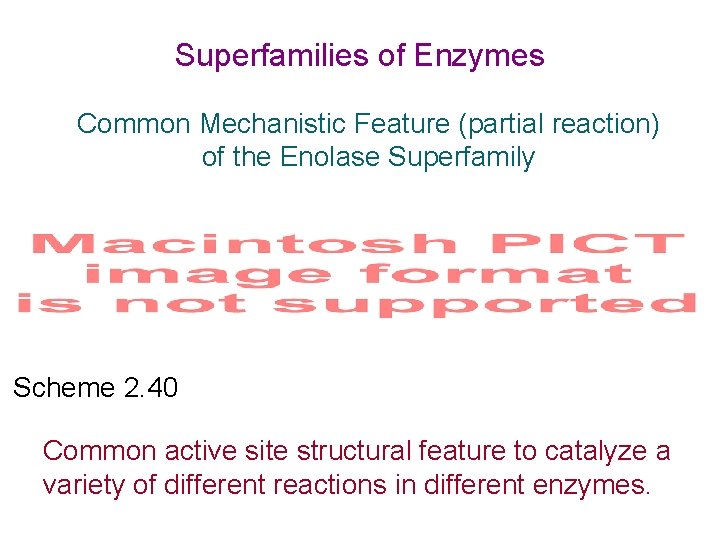
Superfamilies of Enzymes Common Mechanistic Feature (partial reaction) of the Enolase Superfamily Scheme 2. 40 Common active site structural feature to catalyze a variety of different reactions in different enzymes.

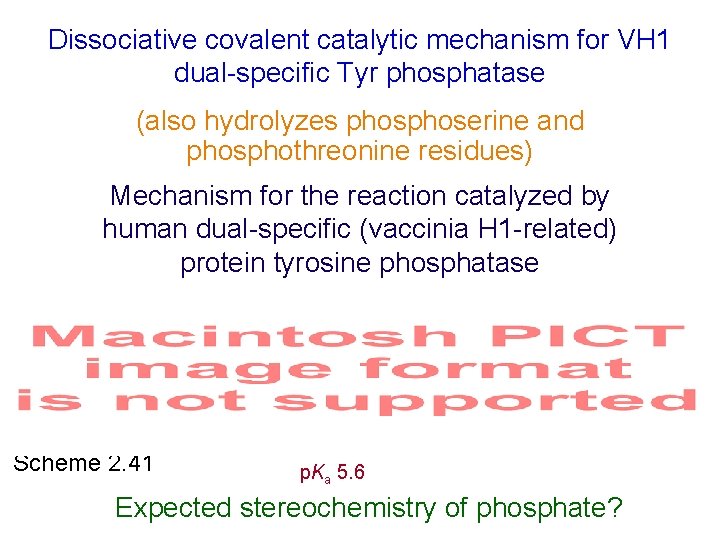
Dissociative covalent catalytic mechanism for VH 1 dual-specific Tyr phosphatase (also hydrolyzes phoserine and phosphothreonine residues) Mechanism for the reaction catalyzed by human dual-specific (vaccinia H 1 -related) protein tyrosine phosphatase Scheme 2. 41 p. Ka 5. 6 Expected stereochemistry of phosphate?

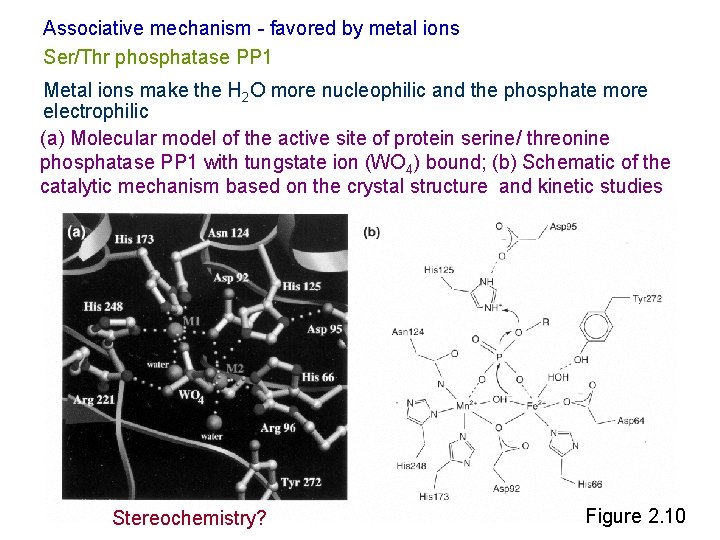
Associative mechanism - favored by metal ions Ser/Thr phosphatase PP 1 Metal ions make the H 2 O more nucleophilic and the phosphate more electrophilic (a) Molecular model of the active site of protein serine/ threonine phosphatase PP 1 with tungstate ion (WO 4) bound; (b) Schematic of the catalytic mechanism based on the crystal structure and kinetic studies Stereochemistry? Figure 2. 10

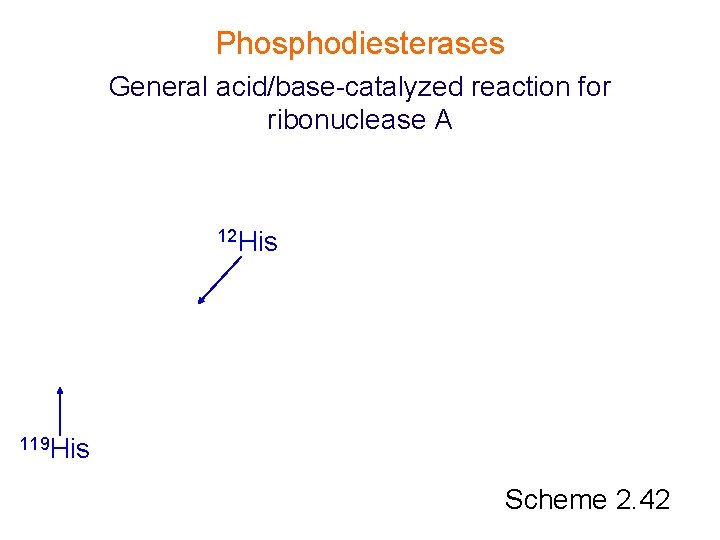
Phosphodiesterases General acid/base-catalyzed reaction for ribonuclease A 12 His 119 His Scheme 2. 42

Kinases • Transfer the -phosphoryl group of nucleoside triphosphates (originally only ATP) to an acceptor • Now generalized to reactions at the -, or -position of any nucleoside triphosphate

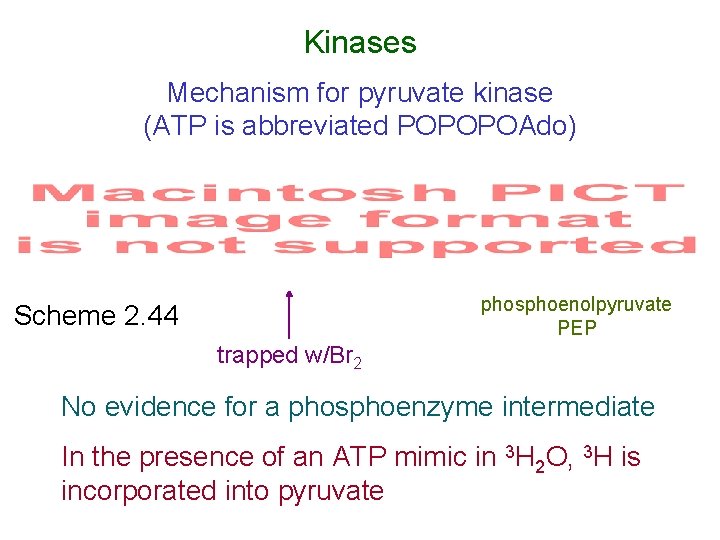
Kinases Mechanism for pyruvate kinase (ATP is abbreviated POPOPOAdo) phosphoenolpyruvate PEP Scheme 2. 44 trapped w/Br 2 No evidence for a phosphoenzyme intermediate In the presence of an ATP mimic in 3 H 2 O, 3 H is incorporated into pyruvate

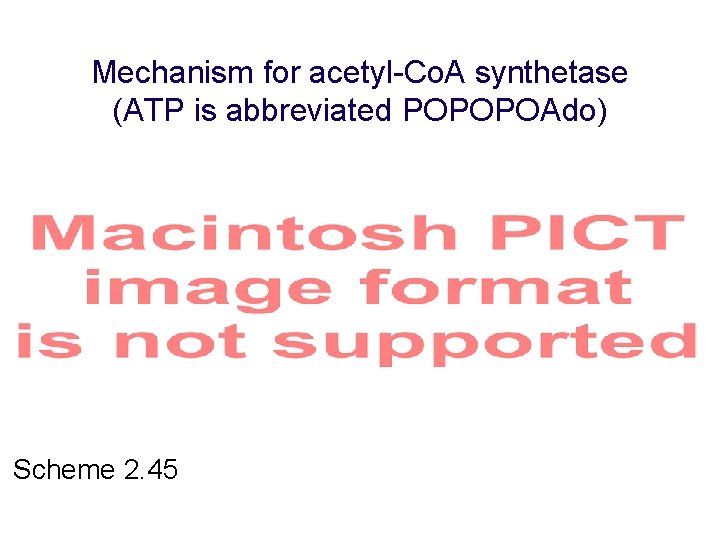
Mechanism for acetyl-Co. A synthetase (ATP is abbreviated POPOPOAdo) Scheme 2. 45