The neutral model approach Stephen P Hubbell 1942

The neutral model approach Stephen P. Hubbell (1942 - Motoo Kimura (1924 -1994)
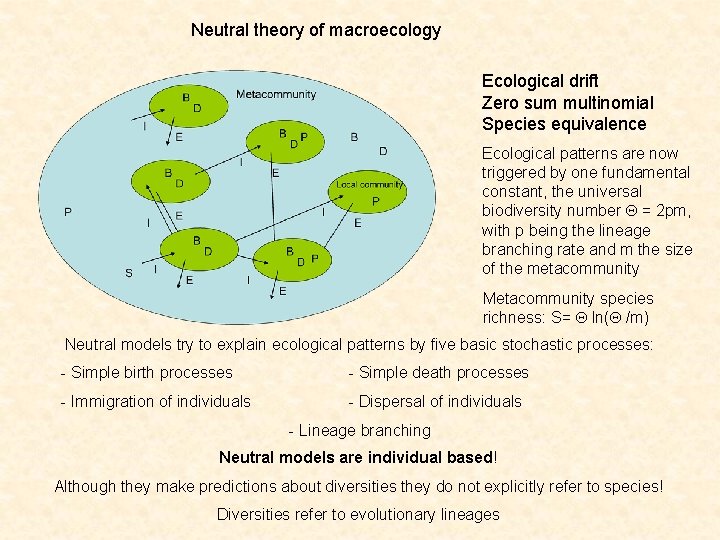
Neutral theory of macroecology Ecological drift Zero sum multinomial Species equivalence Ecological patterns are now triggered by one fundamental constant, the universal biodiversity number Q = 2 pm, with p being the lineage branching rate and m the size of the metacommunity Metacommunity species richness: S= Q ln(Q /m) Neutral models try to explain ecological patterns by five basic stochastic processes: - Simple birth processes - Simple death processes - Immigration of individuals - Dispersal of individuals - Lineage branching Neutral models are individual based! Although they make predictions about diversities they do not explicitly refer to species! Diversities refer to evolutionary lineages
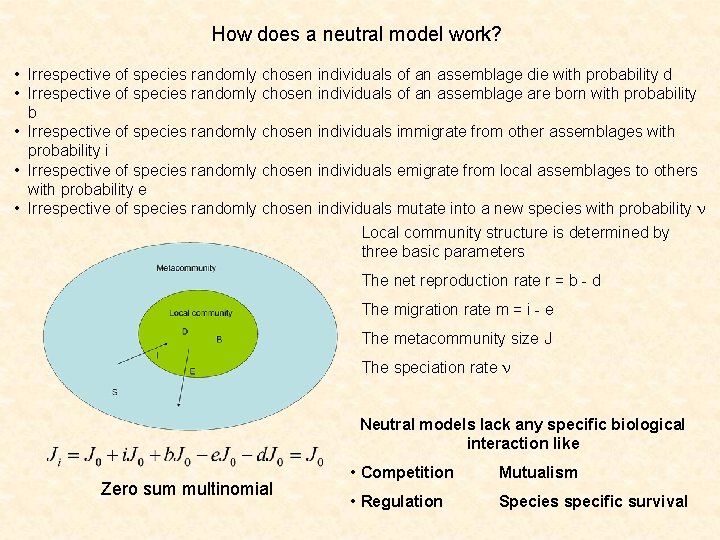
How does a neutral model work? • Irrespective of species randomly chosen individuals of an assemblage die with probability d • Irrespective of species randomly chosen individuals of an assemblage are born with probability b • Irrespective of species randomly chosen individuals immigrate from other assemblages with probability i • Irrespective of species randomly chosen individuals emigrate from local assemblages to others with probability e • Irrespective of species randomly chosen individuals mutate into a new species with probability n Local community structure is determined by three basic parameters The net reproduction rate r = b - d The migration rate m = i - e The metacommunity size J The speciation rate n Neutral models lack any specific biological interaction like Zero sum multinomial • Competition Mutualism • Regulation Species specific survival

The speciation modes 5 patches with individuals of different lineages Point mutation I I I Peripheral isolate I I I Fission track I I I I One individual transforms into a A randomly chosen part of one new lineage with probability n lineage of a local unity lineage of the metacommunity transforms into a new lineage with probability n After many time steps an equilibrium is established Analytical solution exists Fundamental biodiversity number Q a-diversity No analytical solutions yet The metacommuity has a log-series species abundance distribution.
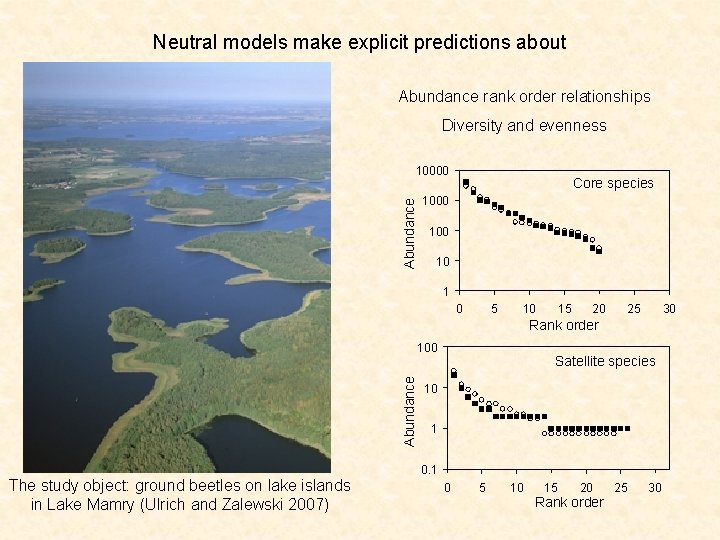
Neutral models make explicit predictions about Abundance rank order relationships Diversity and evenness Abundance 10000 Core species 1000 10 1 Leistus rufomarginatus 0 Photos by Roy Anderson 5 10 20 25 30 Rank order Abundance 100 The study object: ground beetles on lake islands in Lake Mamry (Ulrich and Zalewski 2007) 15 Satellite species 10 1 0 5 10 15 20 Rank order 25 30
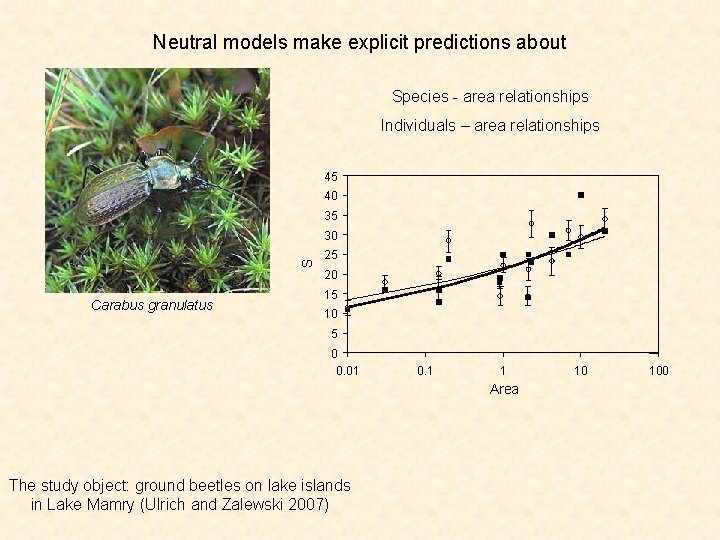
Neutral models make explicit predictions about Species - area relationships Individuals – area relationships 45 40 35 S 30 25 20 Carabus granulatus 15 10 5 0 0. 01 0. 1 1 Area The study object: ground beetles on lake islands in Lake Mamry (Ulrich and Zalewski 2007) 10 100
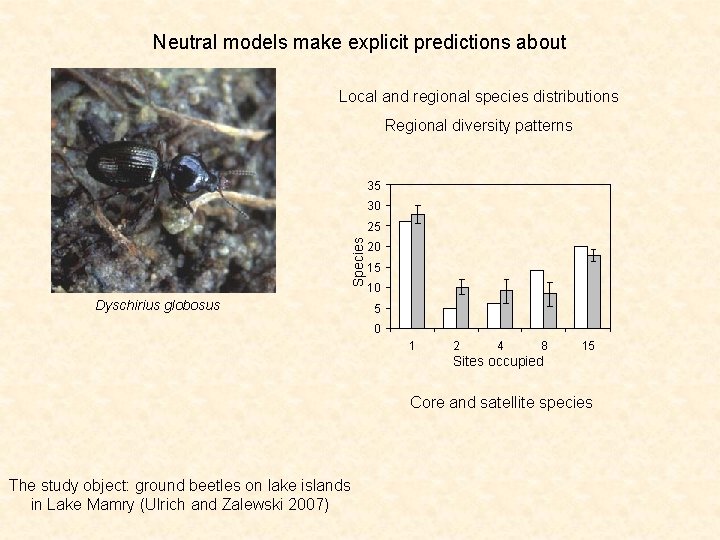
Neutral models make explicit predictions about Local and regional species distributions Regional diversity patterns 35 30 Species 25 Dyschirius globosus 20 15 10 5 0 1 2 4 8 15 Sites occupied Core and satellite species The study object: ground beetles on lake islands in Lake Mamry (Ulrich and Zalewski 2007)
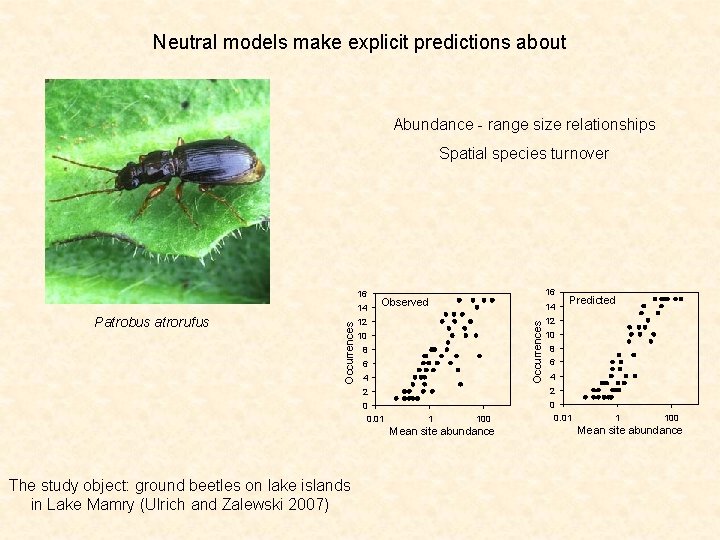
Neutral models make explicit predictions about Abundance - range size relationships Spatial species turnover 16 14 12 Occurrences Patrobus atrorufus Occurrences 14 16 Observed 10 8 6 4 12 10 8 6 4 2 2 0 0 0. 01 1 100 Mean site abundance The study object: ground beetles on lake islands in Lake Mamry (Ulrich and Zalewski 2007) Predicted 1 100 Mean site abundance
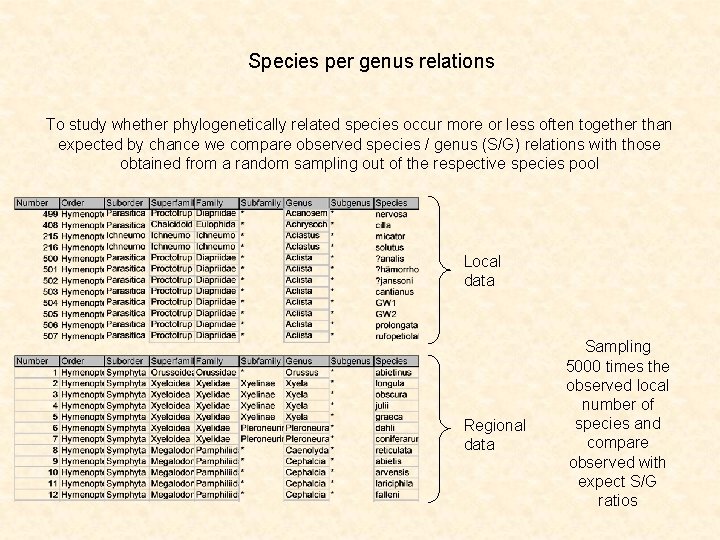
Species per genus relations To study whether phylogenetically related species occur more or less often together than expected by chance we compare observed species / genus (S/G) relations with those obtained from a random sampling out of the respective species pool Local data Regional data Sampling 5000 times the observed local number of species and compare observed with expect S/G ratios
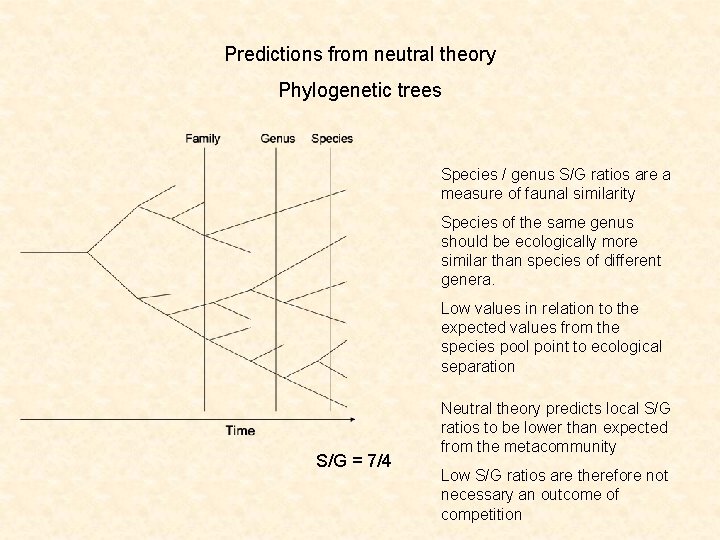
Predictions from neutral theory Phylogenetic trees Species / genus S/G ratios are a measure of faunal similarity Species of the same genus should be ecologically more similar than species of different genera. Low values in relation to the expected values from the species pool point to ecological separation S/G = 7/4 Neutral theory predicts local S/G ratios to be lower than expected from the metacommunity Low S/G ratios are therefore not necessary an outcome of competition
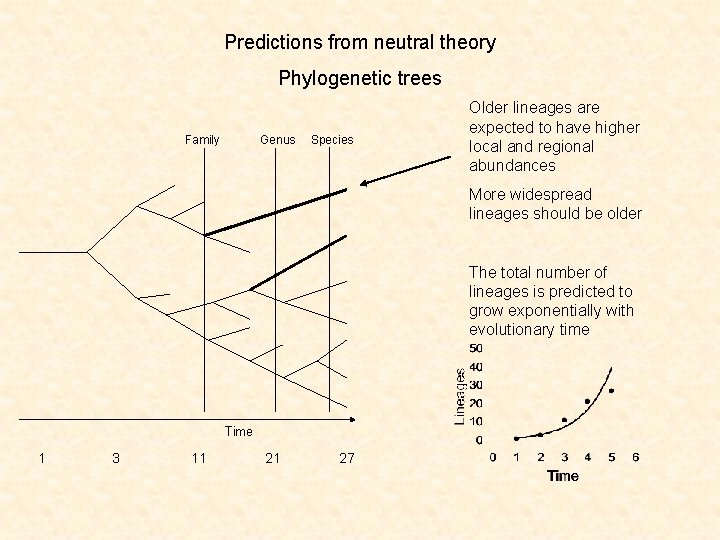
Predictions from neutral theory Phylogenetic trees Family Genus Species Older lineages are expected to have higher local and regional abundances More widespread lineages should be older The total number of lineages is predicted to grow exponentially with evolutionary time Time 1 3 11 21 27
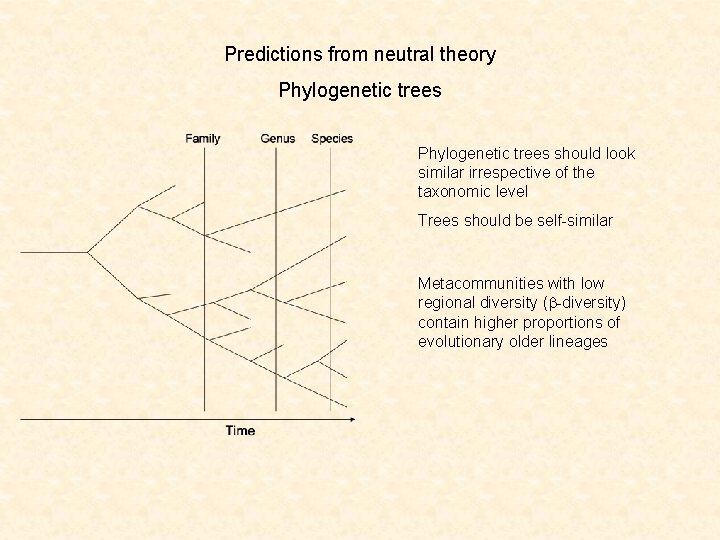
Predictions from neutral theory Phylogenetic trees should look similar irrespective of the taxonomic level Trees should be self-similar Metacommunities with low regional diversity (b-diversity) contain higher proportions of evolutionary older lineages

Today’s reading Neutral theory: http: //en. wikipedia. org/wiki/Unified_neutral_theory_of_biodiversity Null and neutral models: www. uvm. edu/~ngotelli/manuscriptpdfs/gotelli_mcgill_ecography. pdf Neutrality: www. zoology. ufl. edu/rdholt/holtpublications/189. pdf
- Slides: 13