The MORPH Algorithm MORPH MOdule guided Ranking of











- Slides: 45

The MORPH Algorithm MORPH = MOdule guided Ranking of candidate Pat. Hway genes high throughput data Slides: Rachel E. Bell, June 2013

Motivation Challenges in studying biological pathways • Identify missing pathway members • Information gaps on participating genes: a) e. g. nature of interactions between metabolites and gene expression b) understanding control mechanisms, feedback, cross-talk • Many genes in genome(s) have unknown function

Biological Pathways: Overview What is a pathway? A series of interactions between genes (proteins) involved in performing a certain biological function Cell input = extracellular/ endogenous: e. g. : stress, changes in PH, UV exposure, nutrients Cell output = response: e. g. : transcription of genes, sucrose degradation

MORPH Algorithm: Overview ALG INPUT High throughput data of gene expression, networks and biological pathways ORI THM Machine learning and validation methods OUTPUT Predict genes involved in biological pathways

Other methods for functional prediction Coexpression-based methods (& possibly pathways) e. g. : ACT, Gene. Cat, ATED-II, Map. Man Assumptions: 1) Similar expression patterns -> similar function or regulation 2) Pathway genes -> coordinated expression Network-based methods (& gene expression) e. g: Markov random field (MRF) models , k-nearest neighbours (k-NN), ADOMETA: coexpression, phylogeny, clustering on chrom. , metabolic networks Assumption: Closer nodes -> common functions

Introduction: MORPH Algorithm MORPH uses pathway information, gene expression data and network information Compared to other methods, MORPH: • offers robustness (performs well on many pathways) • increases networks coverage • applied to different organisms

Talk outline 1. MORPH input types: (a) gene expression data, (b) pathways and (c) networks 2. Types of clustering (modules) methods 3. The MORPH algorithm and validation 4. Results 5. Comparison to other methods 6. Summary

MORPH Introduction Arabidopsis Thaliana Solanum Lycopersicum (Tomato) MORPH was developed on 2 model organisms

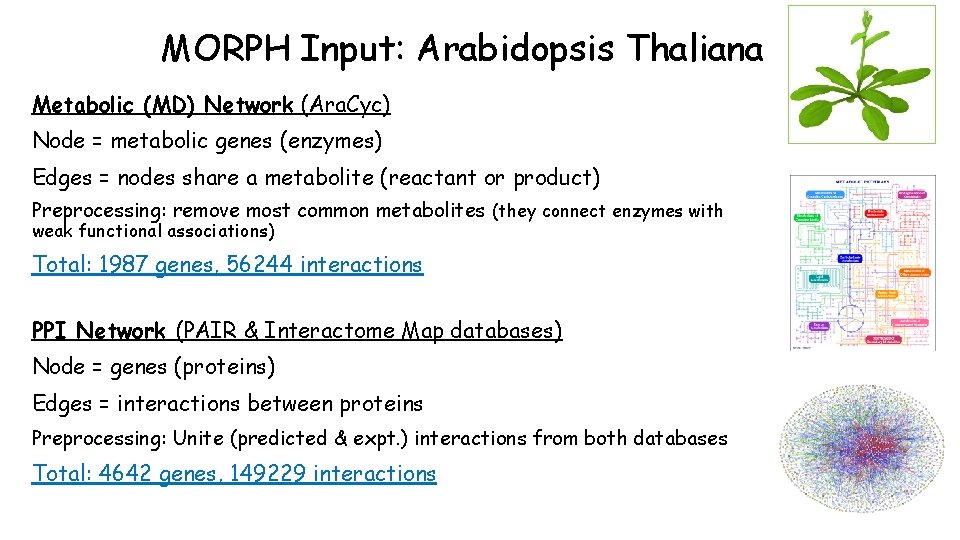
MORPH Input: Arabidopsis Thaliana Pathways: 66 Ara. Cyc, 164 Map. Man Preprocessing: filter pathways with <10 genes with expression data Total 230 pathways, 2 sets Gene Expression datasets: seedlings, tissues (leaves, roots, flowers, seeds), seed developmental stages, DS 1 Preprocessing: filter low variance and detection call, average replicates, normalize to controls, standardize experiments Total 216 GE profiles, 4 datasets, ~12500 genes

MORPH Input: Arabidopsis Thaliana Metabolic (MD) Network (Ara. Cyc) Node = metabolic genes (enzymes) Edges = nodes share a metabolite (reactant or product) Preprocessing: remove most common metabolites (they connect enzymes with weak functional associations) Total: 1987 genes, 56244 interactions PPI Network (PAIR & Interactome Map databases) Node = genes (proteins) Edges = interactions between proteins Preprocessing: Unite (predicted & expt. ) interactions from both databases Total: 4642 genes, 149229 interactions

Talk outline 1. MORPH input types: (a) gene expression data, (b) pathways and (c) networks 2. Types of clustering (modules) methods 3. The MORPH algorithm and validation 4. Results 5. Comparison to other methods 6. Summary

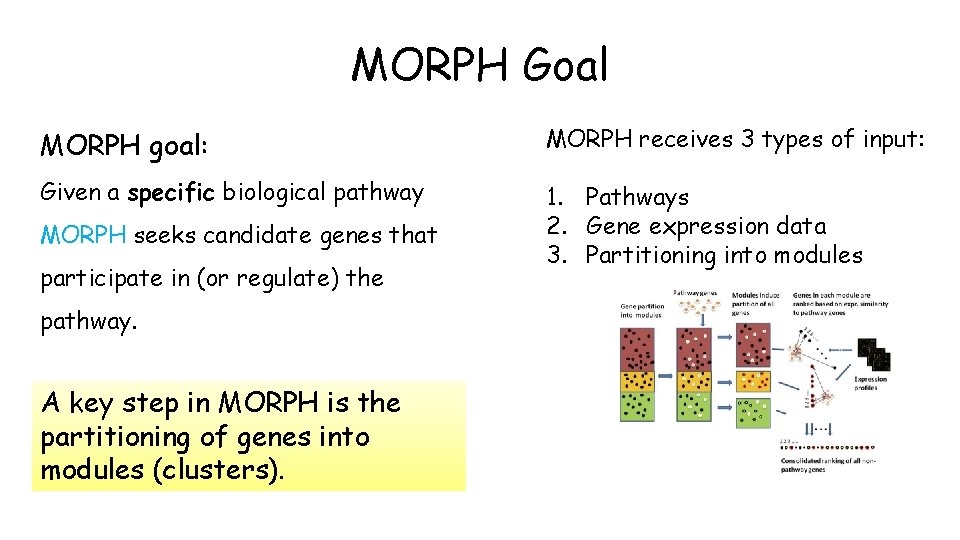
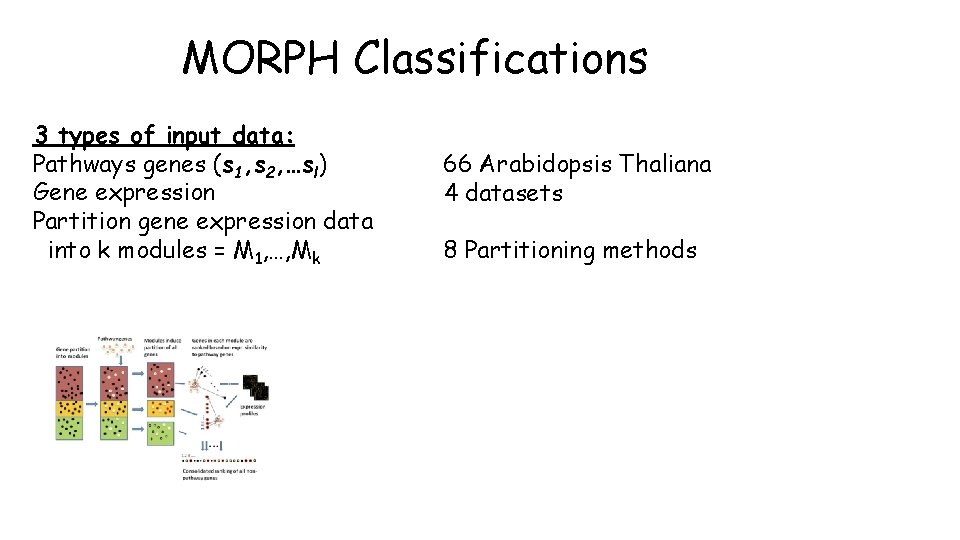
MORPH Goal MORPH goal: MORPH receives 3 types of input: Given a specific biological pathway 1. Pathways 2. Gene expression data 3. Partitioning into modules MORPH seeks candidate genes that participate in (or regulate) the pathway. A key step in MORPH is the partitioning of genes into modules (clusters).

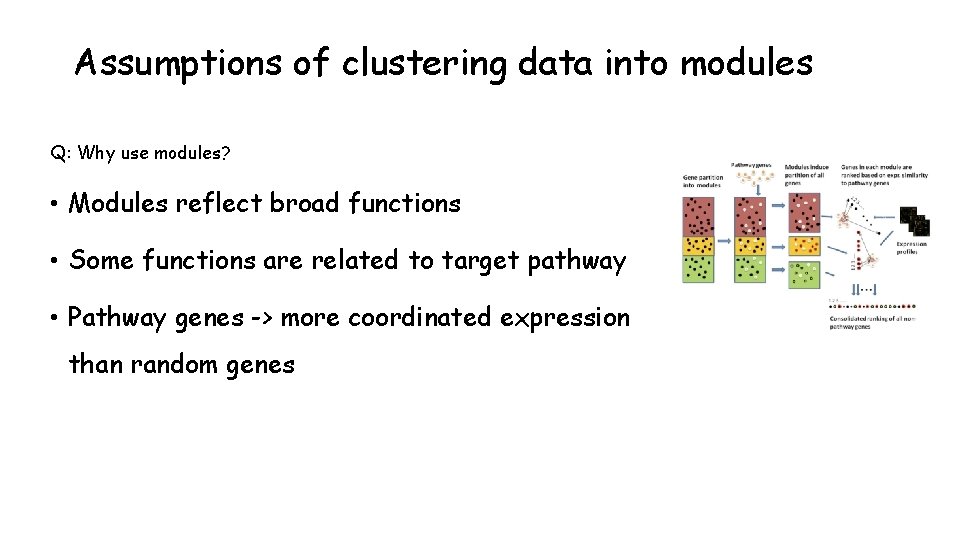
Assumptions of clustering data into modules Q: Why use modules? • Modules reflect broad functions • Some functions are related to target pathway • Pathway genes -> more coordinated expression than random genes

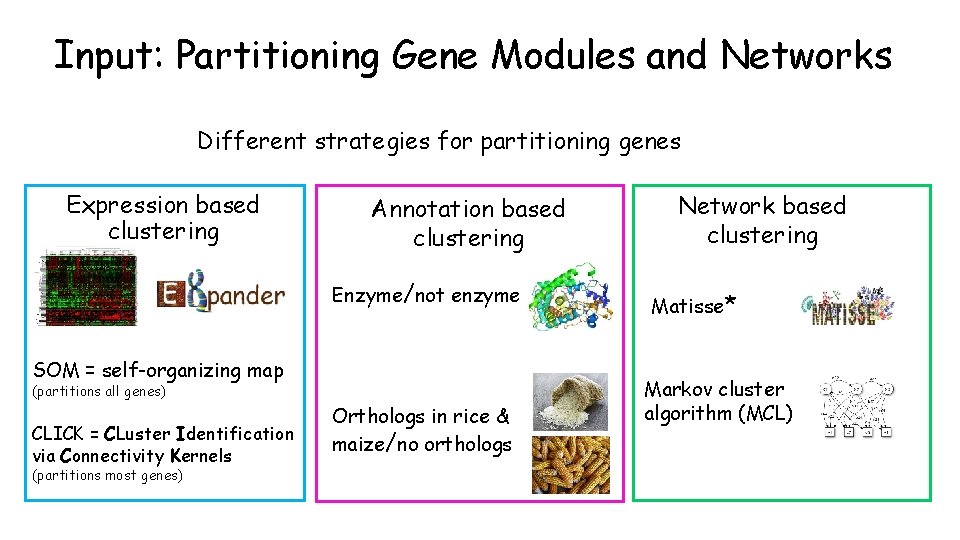
Input: Partitioning Gene Modules and Networks Different strategies for partitioning genes Expression based clustering Annotation based clustering Enzyme/not enzyme SOM = self-organizing map (partitions all genes) CLICK = CLuster Identification via Connectivity Kernels (partitions most genes) Orthologs in rice & maize/no orthologs Network based clustering Matisse* Markov cluster algorithm (MCL)

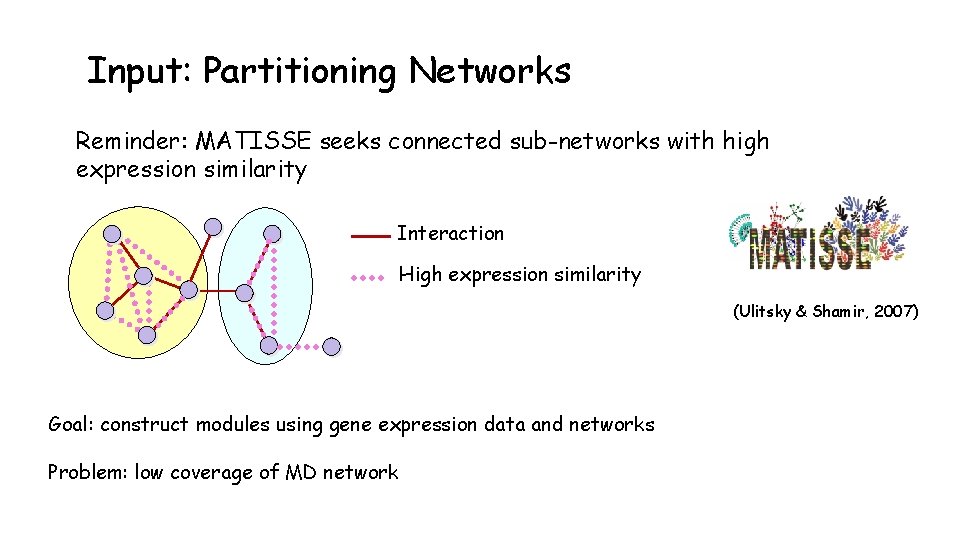
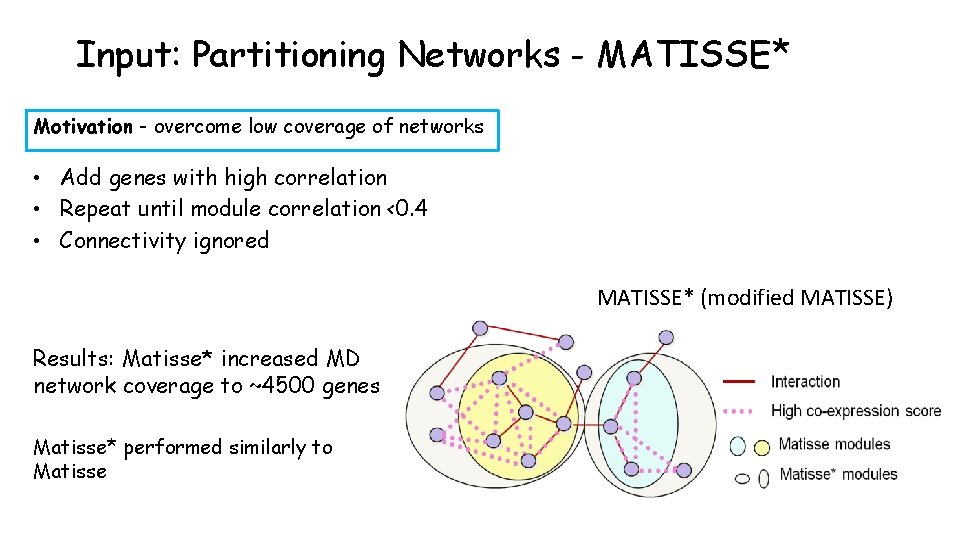
Input: Partitioning Networks Reminder: MATISSE seeks connected sub-networks with high expression similarity Interaction High expression similarity (Ulitsky & Shamir, 2007) Goal: construct modules using gene expression data and networks Problem: low coverage of MD network

Input: Partitioning Networks - MATISSE* Motivation - overcome low coverage of networks • Add genes with high correlation • Repeat until module correlation <0. 4 • Connectivity ignored MATISSE* (modified MATISSE) Results: Matisse* increased MD network coverage to ~4500 genes Matisse* performed similarly to Matisse

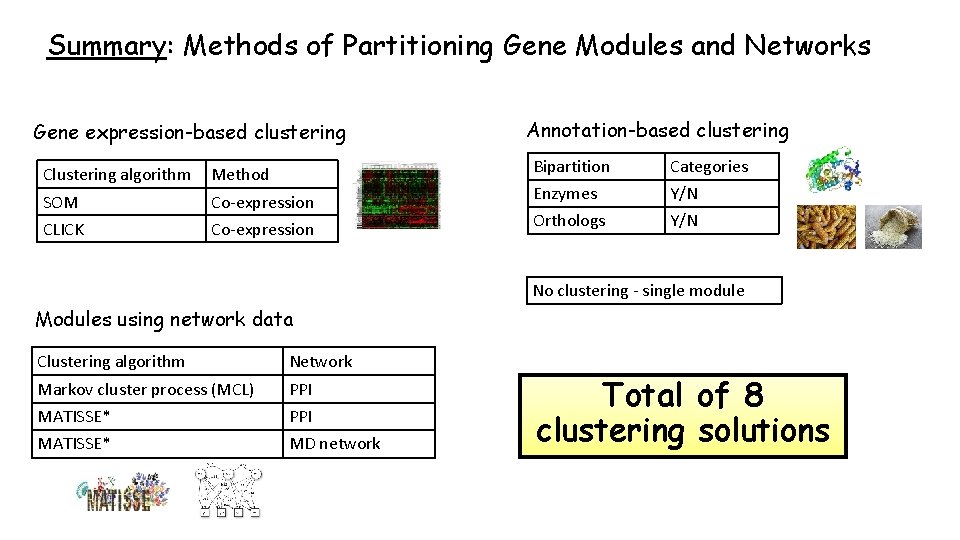
Summary: Methods of Partitioning Gene Modules and Networks Gene expression-based clustering Clustering algorithm Method SOM CLICK Co-expression Annotation-based clustering Bipartition Enzymes Orthologs Categories Y/N No clustering - single module Modules using network data Clustering algorithm Network Markov cluster process (MCL) PPI MATISSE* PPI MD network Total of 8 clustering solutions

Talk outline 1. MORPH input types: (a) gene expression data, (b) pathways and (c) networks 2. Types of clustering (modules) methods 3. The MORPH algorithm and validation 4. Results 5. Comparison to other methods 6. Summary

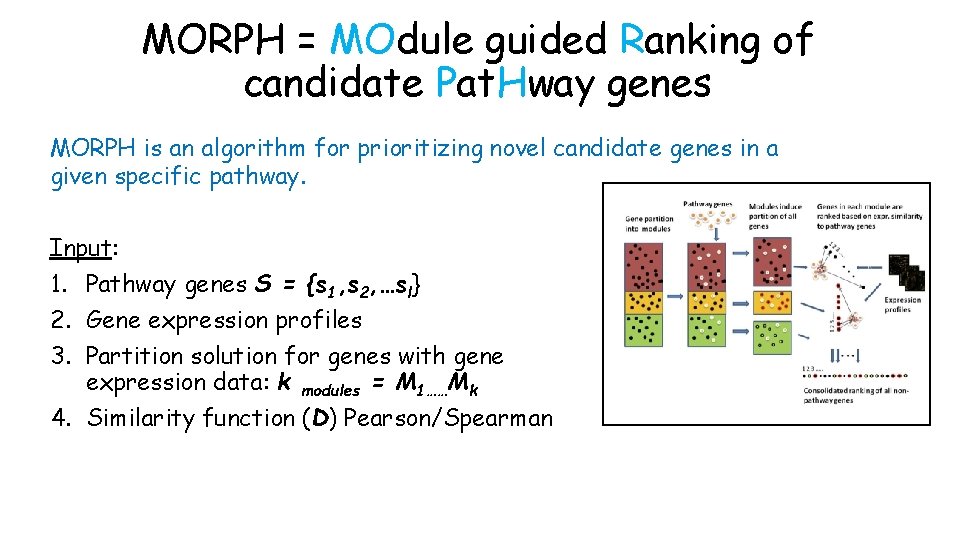
MORPH = MOdule guided Ranking of candidate Pat. Hway genes MORPH is an algorithm for prioritizing novel candidate genes in a given specific pathway. Input: 1. Pathway genes S = {s 1, s 2, …sl} 2. Gene expression profiles 3. Partition solution for genes with gene expression data: k modules = M 1……Mk 4. Similarity function (D) Pearson/Spearman

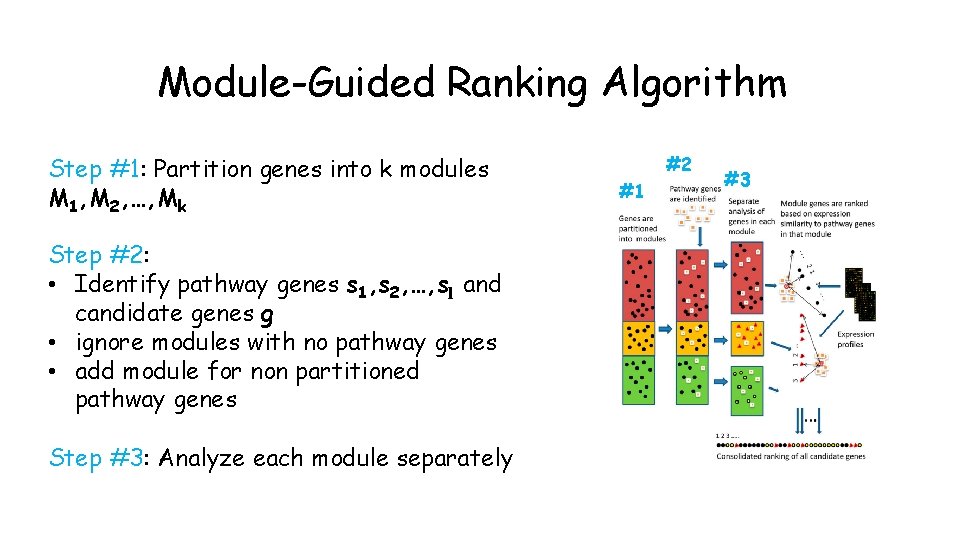
Module-Guided Ranking Algorithm Step #1: Partition genes into k modules M 1, M 2, …, Mk Step #2: • Identify pathway genes s 1, s 2, …, sl and candidate genes g • ignore modules with no pathway genes • add module for non partitioned pathway genes Step #3: Analyze each module separately #2 #1 #3

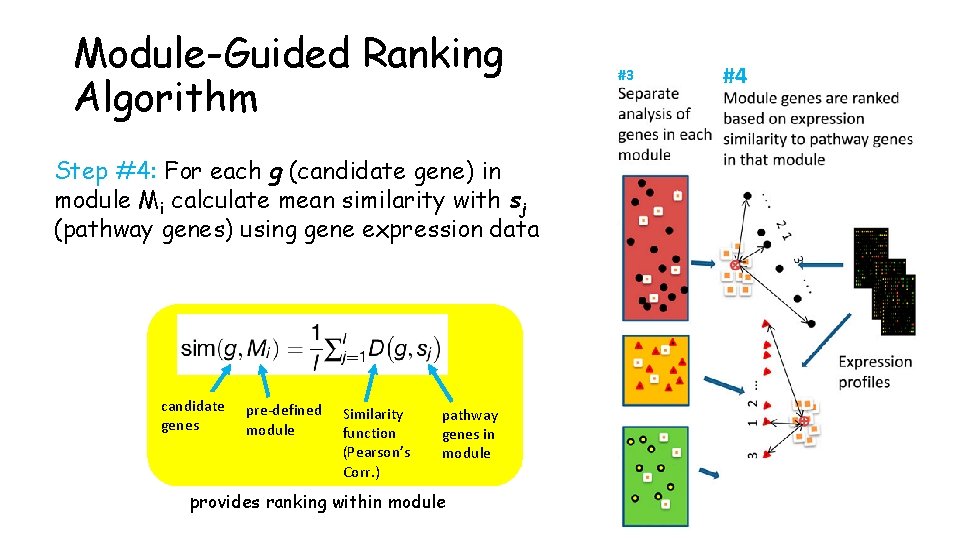
Module-Guided Ranking Algorithm Step #4: For each g (candidate gene) in module Mi calculate mean similarity with sj (pathway genes) using gene expression data candidate genes pre-defined module Similarity function (Pearson’s Corr. ) pathway genes in module provides ranking within module #3 #4

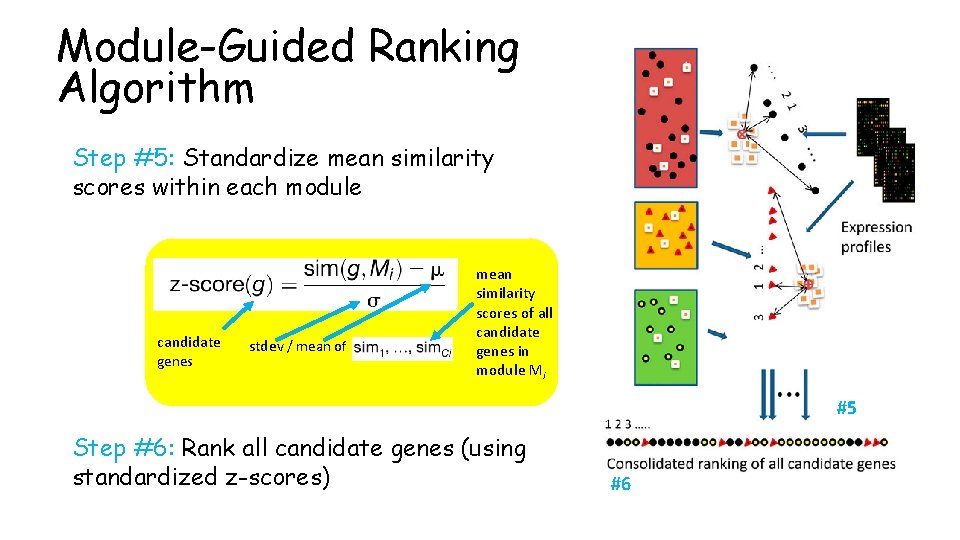
Module-Guided Ranking Algorithm Step #5: Standardize mean similarity scores within each module candidate genes stdev / mean of mean similarity scores of all candidate genes in module Mi #5 Step #6: Rank all candidate genes (using standardized z-scores) #6

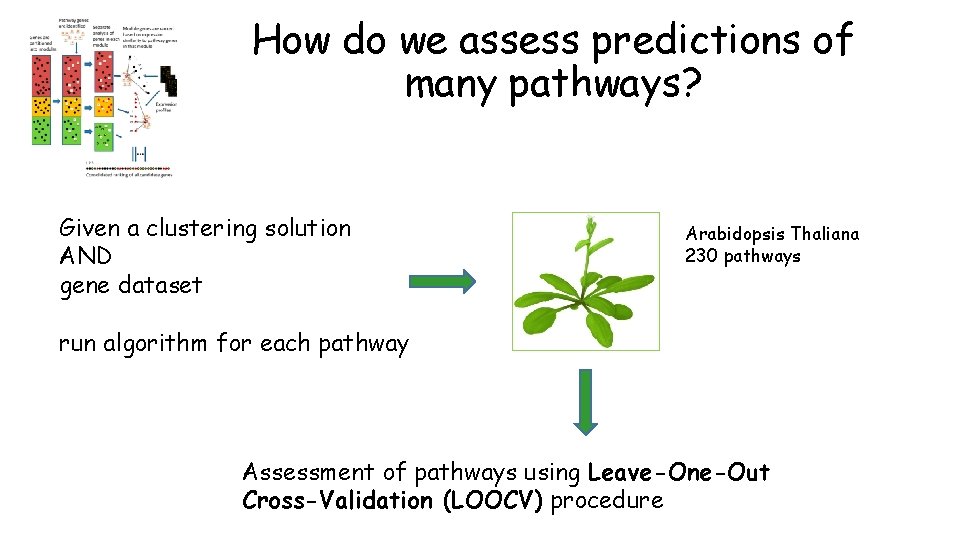
How do we assess predictions of many pathways? Given a clustering solution AND gene dataset Arabidopsis Thaliana 230 pathways run algorithm for each pathway Assessment of pathways using Leave-One-Out Cross-Validation (LOOCV) procedure

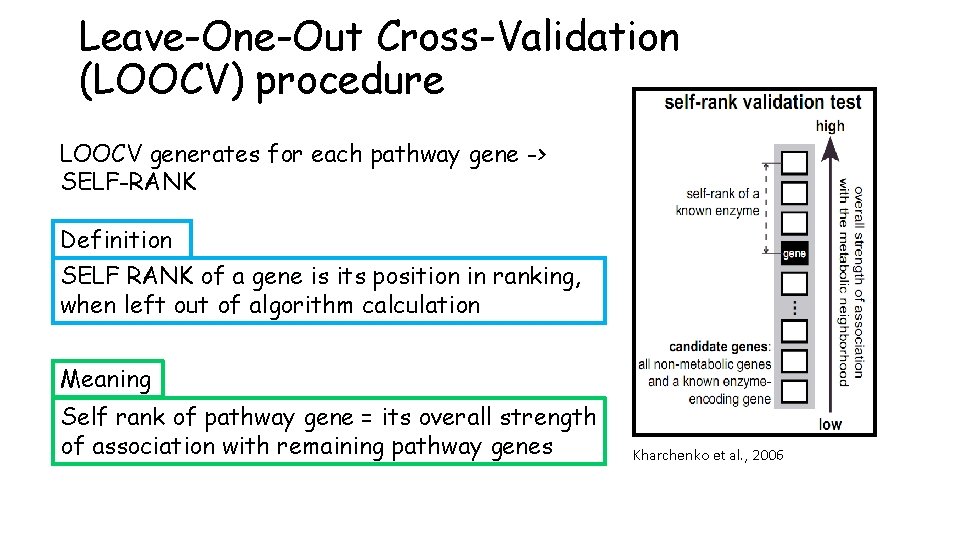
Leave-One-Out Cross-Validation (LOOCV) procedure LOOCV generates for each pathway gene -> SELF-RANK Definition SELF RANK of a gene is its position in ranking, when left out of algorithm calculation Meaning Self rank of pathway gene = its overall strength of association with remaining pathway genes Kharchenko et al. , 2006

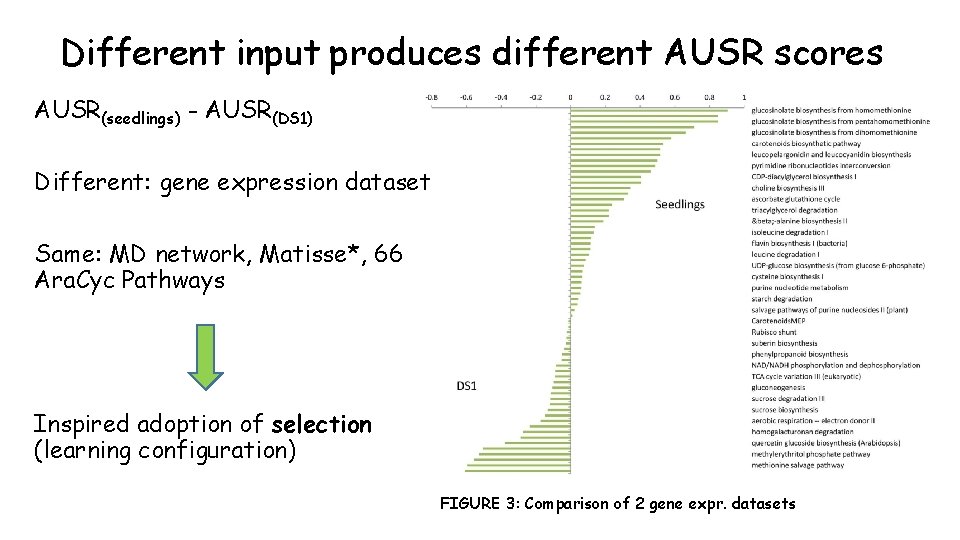
Self-Rank Curve: AUSR score LOOCV procedure For each pathway S: 1. Remove one gene (v) -> S{v} 2. Consider S{v} = test set 3. Generate ranking of v using S{v} 4. Repeat for every v • • Calculate self-rank for all v in S Create self-rank plot Self-rank threshold of k=1. . 1000 Calculate area under self-rank curve (AUSR) (Random gene set of size 13 genes) AUSR score assesses pathway solutions (given input combinations – discussed next) (k) Figure 2 Self-Rank plot of the Carotenoid Biosynthetic Pathway contains 13 genes; SOM - clustering solution

Talk outline 1. MORPH input types: (a) gene expression data, (b) pathways and (c) networks 2. Types of clustering (modules) methods 3. The MORPH algorithm and validation 4. Results 5. Comparison to other methods 6. Summary

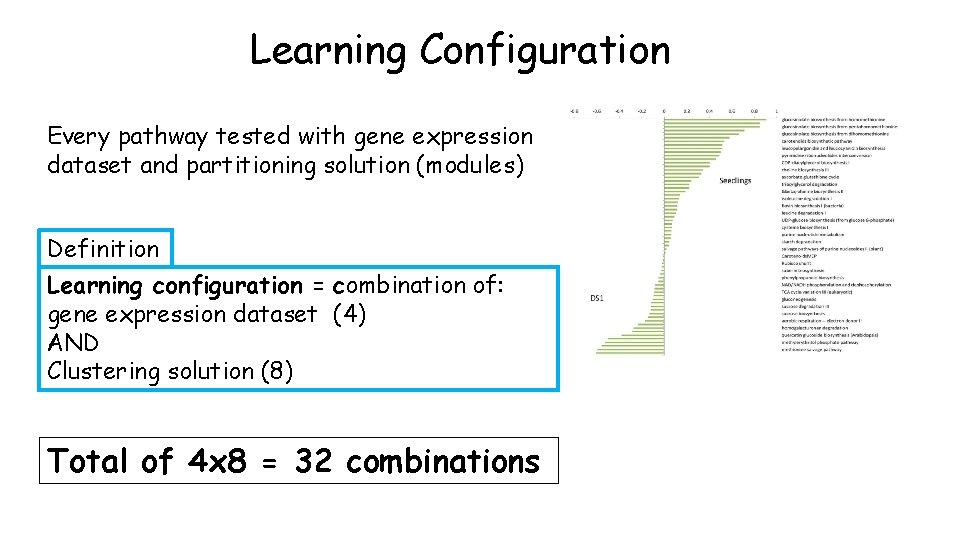
Different input produces different AUSR scores AUSR(seedlings) - AUSR(DS 1) Different: gene expression dataset Same: MD network, Matisse*, 66 Ara. Cyc Pathways Inspired adoption of selection (learning configuration) FIGURE 3: Comparison of 2 gene expr. datasets

Learning Configuration Every pathway tested with gene expression dataset and partitioning solution (modules) Definition Learning configuration = combination of: gene expression dataset (4) AND Clustering solution (8) Total of 4 x 8 = 32 combinations

Machine Learning LOOCV used to select optimal learning configuration (i. e. data set and clustering) for each examined pathway. LOOCV avoids overfitting, since test gene is left out. MORPH applies a selection procedure

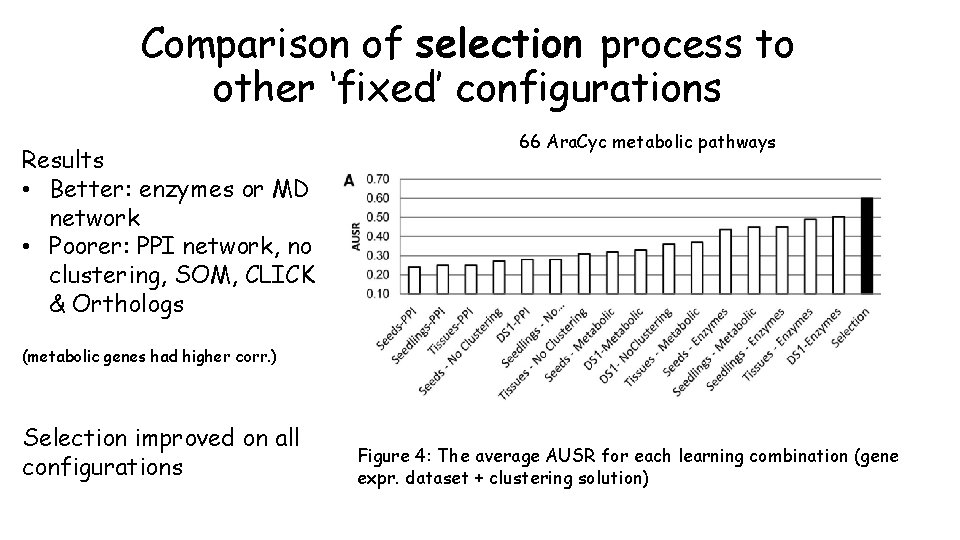
Comparison of selection process to other ‘fixed’ configurations Results • Better: enzymes or MD network • Poorer: PPI network, no clustering, SOM, CLICK & Orthologs 66 Ara. Cyc metabolic pathways (metabolic genes had higher corr. ) Selection improved on all configurations Figure 4: The average AUSR for each learning combination (gene expr. dataset + clustering solution)

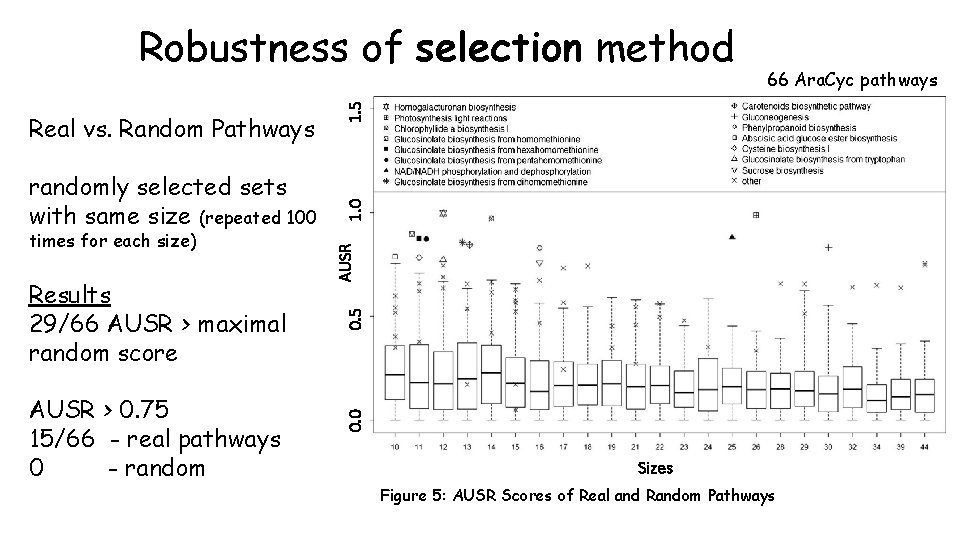
Results 29/66 AUSR > maximal random score AUSR > 0. 75 15/66 - real pathways 0 - random 1. 0 AUSR times for each size) 0. 5 randomly selected sets with same size (repeated 100 0. 0 Real vs. Random Pathways 66 Ara. Cyc pathways 1. 5 Robustness of selection method Sizes Figure 5: AUSR Scores of Real and Random Pathways

Talk outline 1. MORPH input types: (a) gene expression data, (b) pathways and (c) networks 2. Types of clustering (modules) methods 3. The MORPH algorithm and validation 4. Results 5. Comparison to other methods 6. Summary

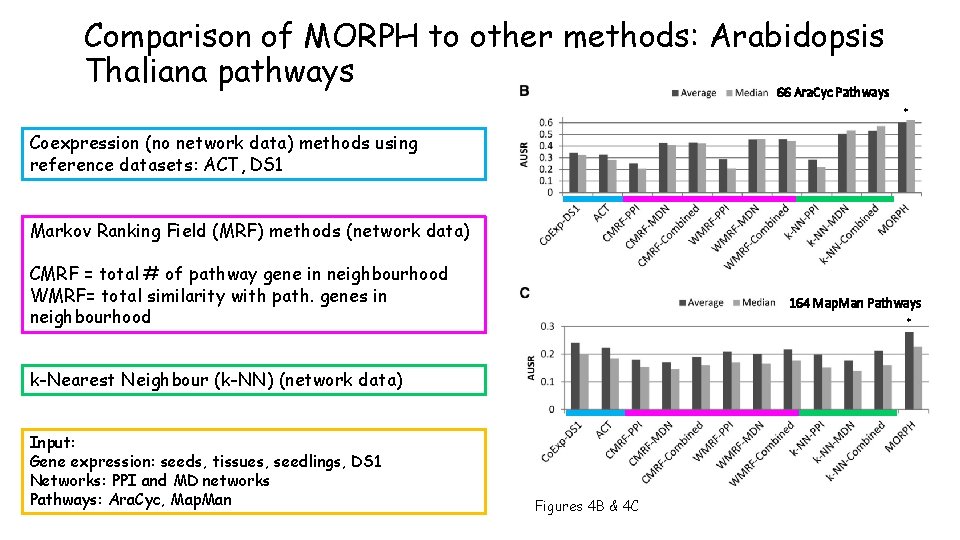
Comparison of MORPH to other methods: Arabidopsis Thaliana pathways 66 Ara. Cyc Pathways * Coexpression (no network data) methods using reference datasets: ACT, DS 1 Markov Ranking Field (MRF) methods (network data) CMRF = total # of pathway gene in neighbourhood WMRF= total similarity with path. genes in neighbourhood 164 Map. Man Pathways * k-Nearest Neighbour (k-NN) (network data) Input: Gene expression: seeds, tissues, seedlings, DS 1 Networks: PPI and MD networks Pathways: Ara. Cyc, Map. Man Figures 4 B & 4 C

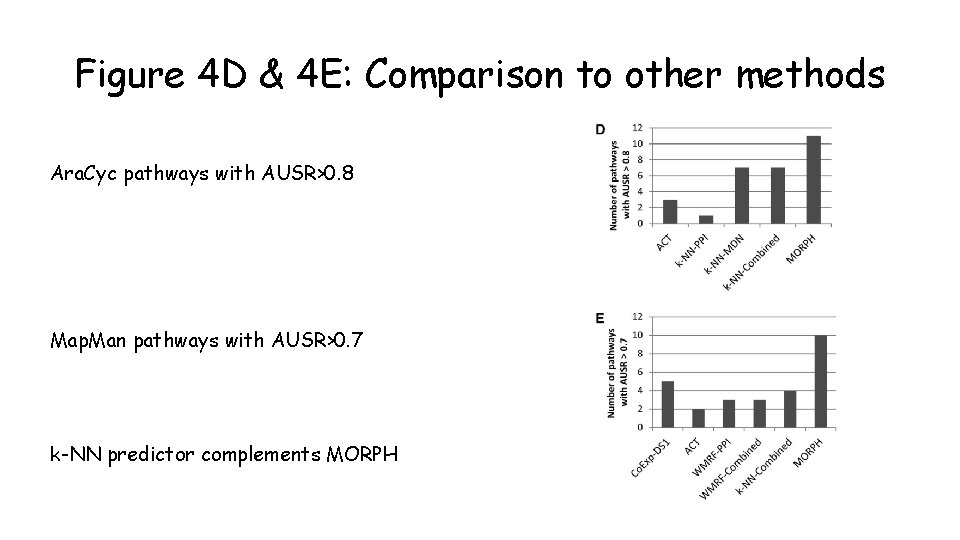
Figure 4 D & 4 E: Comparison to other methods Ara. Cyc pathways with AUSR>0. 8 Map. Man pathways with AUSR>0. 7 k-NN predictor complements MORPH

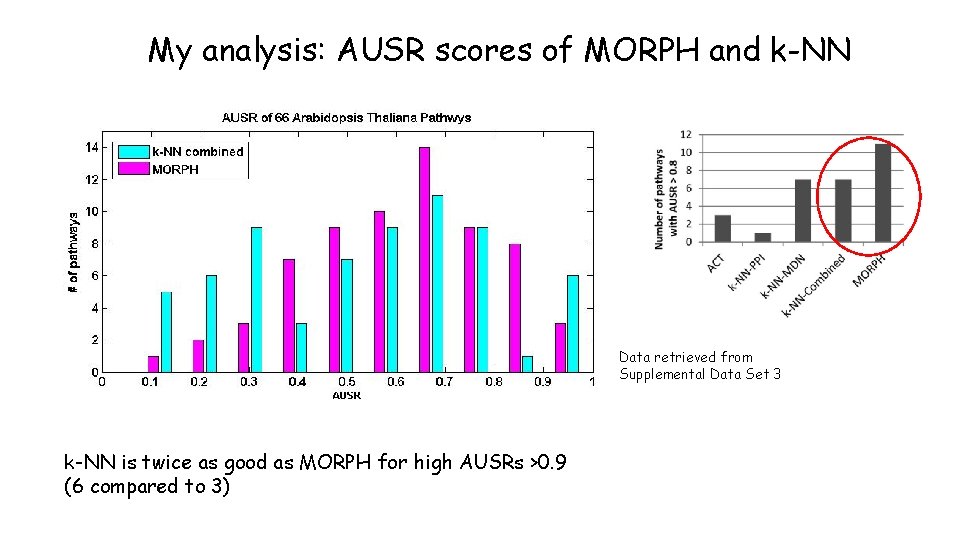
My analysis: AUSR scores of MORPH and k-NN Data retrieved from Supplemental Data Set 3 k-NN is twice as good as MORPH for high AUSRs >0. 9 (6 compared to 3)

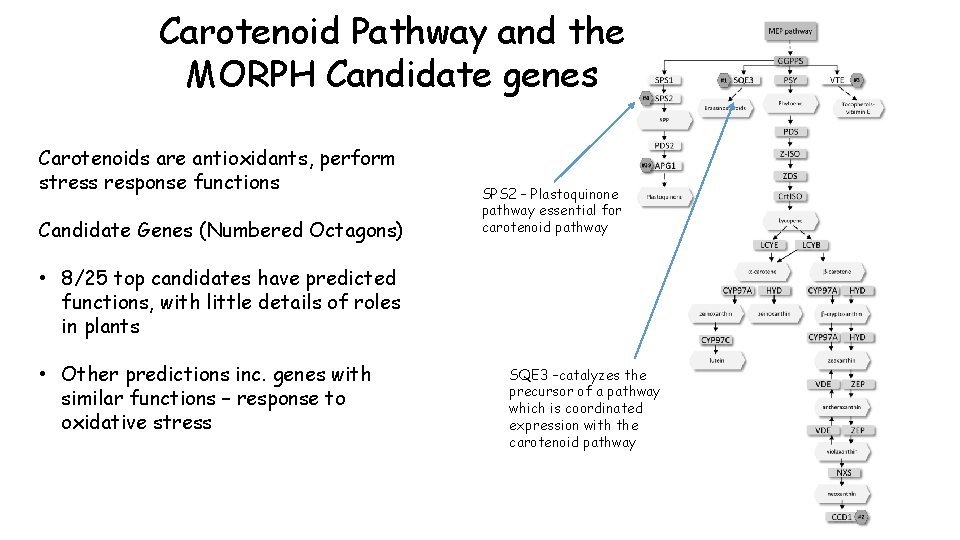
Carotenoid Pathway and the MORPH Candidate genes Carotenoids are antioxidants, perform stress response functions Candidate Genes (Numbered Octagons) SPS 2 – Plastoquinone pathway essential for carotenoid pathway • 8/25 top candidates have predicted functions, with little details of roles in plants • Other predictions inc. genes with similar functions – response to oxidative stress SQE 3 –catalyzes the precursor of a pathway which is coordinated expression with the carotenoid pathway

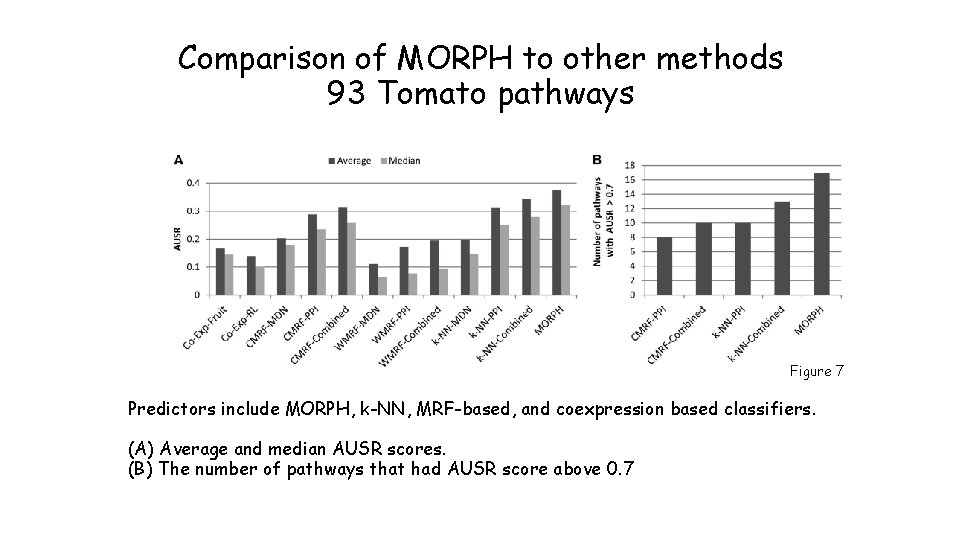
Comparison of MORPH to other methods 93 Tomato pathways Figure 7 Predictors include MORPH, k-NN, MRF-based, and coexpression based classifiers. (A) Average and median AUSR scores. (B) The number of pathways that had AUSR score above 0. 7

Talk outline 1. MORPH input types: (a) gene expression data, (b) pathways and (c) networks 2. Types of clustering (modules) methods 3. The MORPH algorithm and validation 4. Results 5. Comparison to other methods 6. Summary

Summary: Advantages of MORPH 1. Robust – different pathways 2. k-NN consider only genes in the network, MORPH increases network coverage 3. k-NN more dependent on sub-networks diameter (higher diameter lower AUSR), MORPH more robust 4. Self-rank k=1000 threshold for AUSR, ignores poor pathway gene correlations 5. Potential useful predictions

Summary: Drawbacks of MORPH 1. If pathway genes not coherent, better select best/top module(s) than average 2. Dependent on input quality (e. g. Ara. Cyc > Map. Man) 3. Predicts close pathways (drawback/advantage) 4. Requires known pathway info for predictions

Questions?


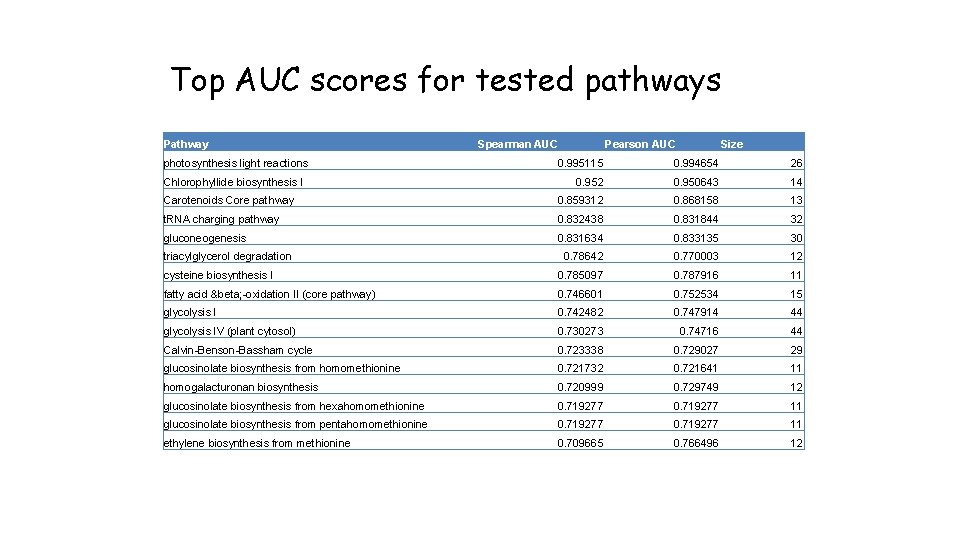
Top AUC scores for tested pathways Pathway photosynthesis light reactions Spearman AUC Pearson AUC Size 0. 995115 0. 994654 26 0. 952 0. 950643 14 Carotenoids Core pathway 0. 859312 0. 868158 13 t. RNA charging pathway 0. 832438 0. 831844 32 gluconeogenesis 0. 831634 0. 833135 30 0. 78642 0. 770003 12 cysteine biosynthesis I 0. 785097 0. 787916 11 fatty acid β -oxidation II (core pathway) 0. 746601 0. 752534 15 glycolysis I 0. 742482 0. 747914 44 glycolysis IV (plant cytosol) 0. 730273 0. 74716 44 Calvin-Benson-Bassham cycle 0. 723338 0. 729027 29 glucosinolate biosynthesis from homomethionine 0. 721732 0. 721641 11 homogalacturonan biosynthesis 0. 720999 0. 729749 12 glucosinolate biosynthesis from hexahomomethionine 0. 719277 11 glucosinolate biosynthesis from pentahomomethionine 0. 719277 11 ethylene biosynthesis from methionine 0. 709665 0. 766496 12 Chlorophyllide biosynthesis I triacylglycerol degradation

MORPH Classifications 3 types of input data: Pathways genes (s 1, s 2, …sl) Gene expression Partition gene expression data into k modules = M 1, …, Mk 66 Arabidopsis Thaliana 4 datasets 8 Partitioning methods
