The MITK Approach Ivo Wolf Marco Nolden Thomas





- Slides: 30

The MITK Approach Ivo Wolf, Marco Nolden, Thomas Böttger, Ingmar Wegner, Max Schöbinger, Daniel Maleike, Mathias Seitel, Urte Rietdorf, Daniel Stein, Michael Hasselberg, Tobias Heimann, Mark Hastenteufel, Hans-Peter Meinzer, Marcus Vetter Div. Medical and Biological Informatics German Cancer Research Center, Heidelberg

Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 2 MITK at a glance MITK = Medical Imaging Interaction Toolkit So … § medical imaging § interactive systems § toolkit, not an application or development environment

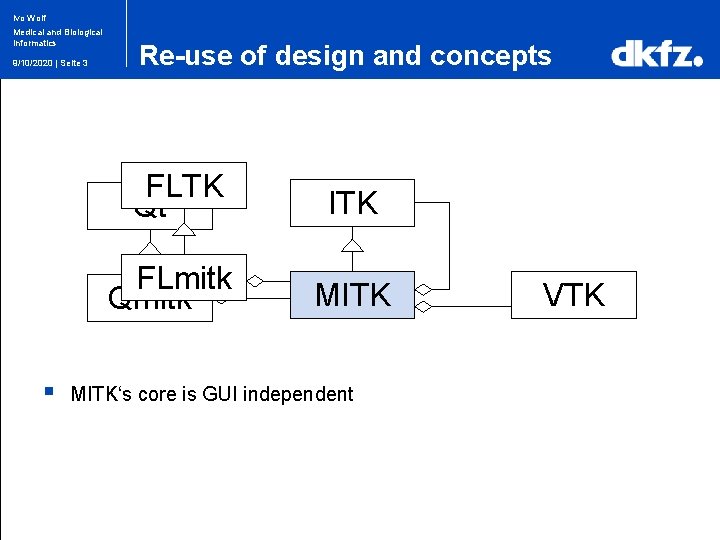
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 3 § Re-use of design and concepts FLTK Qt ITK FLmitk Qmitk MITK‘s core is GUI independent VTK

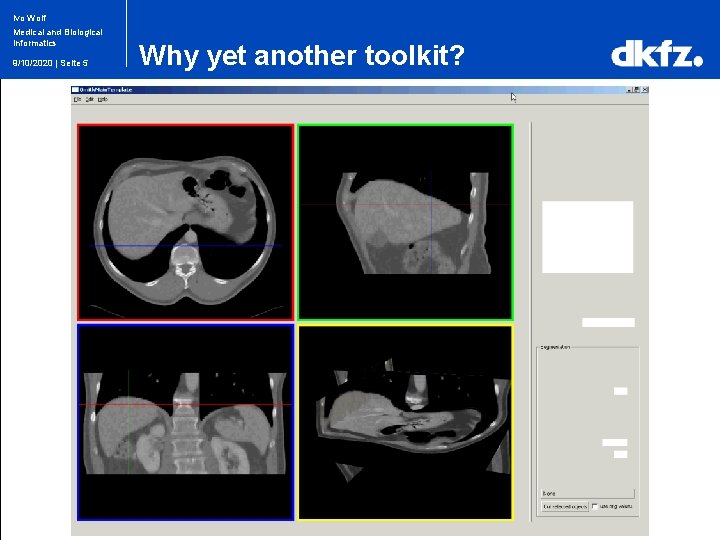
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 4 Why yet another toolkit?

Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 5 Why yet another toolkit?

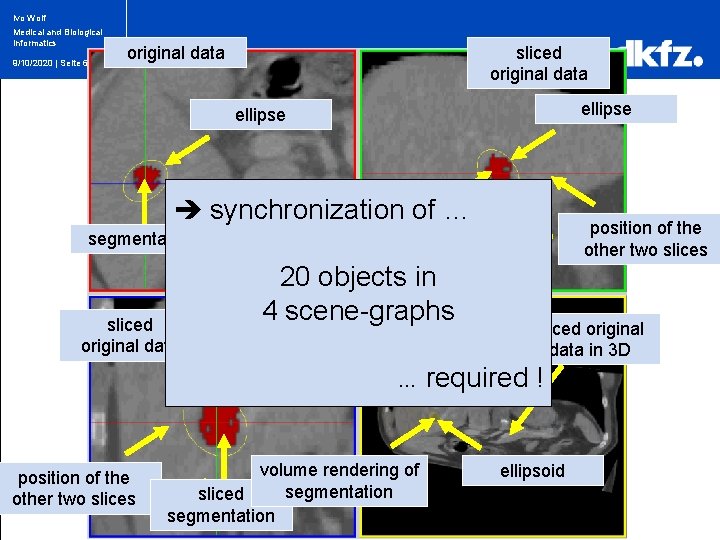
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 6 original data sliced original data ellipse synchronizationsliced of … segmentation sliced original data 20 objects in 4 scene-graphs ellipse … position of the other two slices position of the segmentation other two slices volume rendering of segmentation sliced original data in 3 D required ! ellipsoid

Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 7 Why yet another toolkit? Of course this could be done … … but what when we want to see an additional surface: for the liver boundary 4 more scene objects and the vessels again 4 more scene objects … the segmentation could also be useful for our heart projects … and we want to see segmentations of the ventricels, the annulus and the coronaries and …

Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 8 Why yet another toolkit? We would end up here …

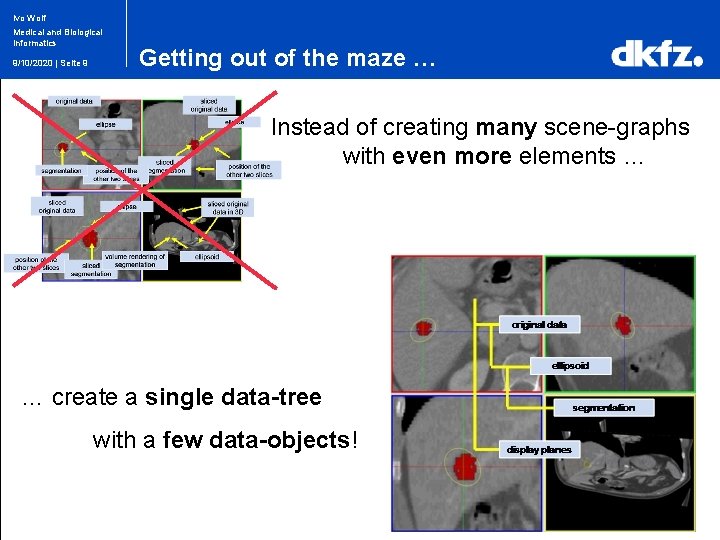
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 9 Getting out of the maze … Instead of creating many scene-graphs with even more elements … … create a single data-tree with a few data-objects!

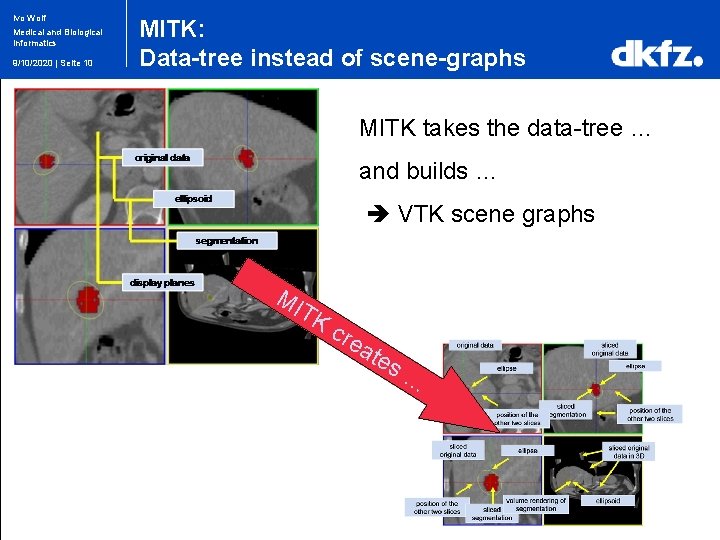
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 10 MITK: Data-tree instead of scene-graphs MITK takes the data-tree … and builds … VTK scene graphs MI TK cre ate s …

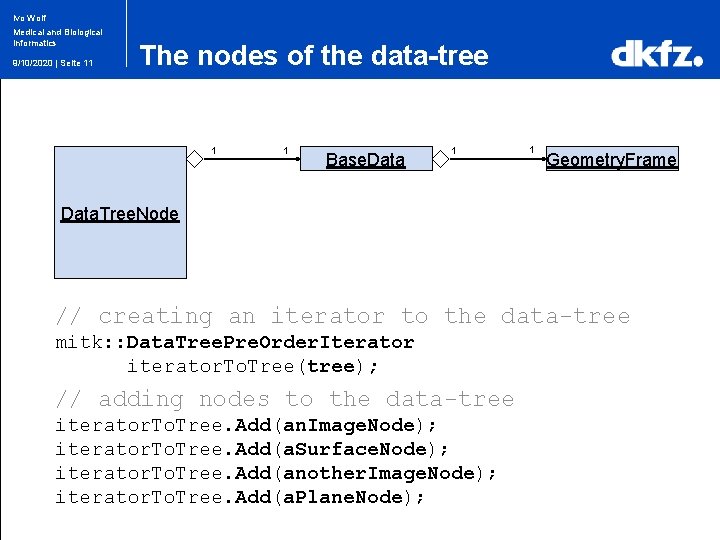
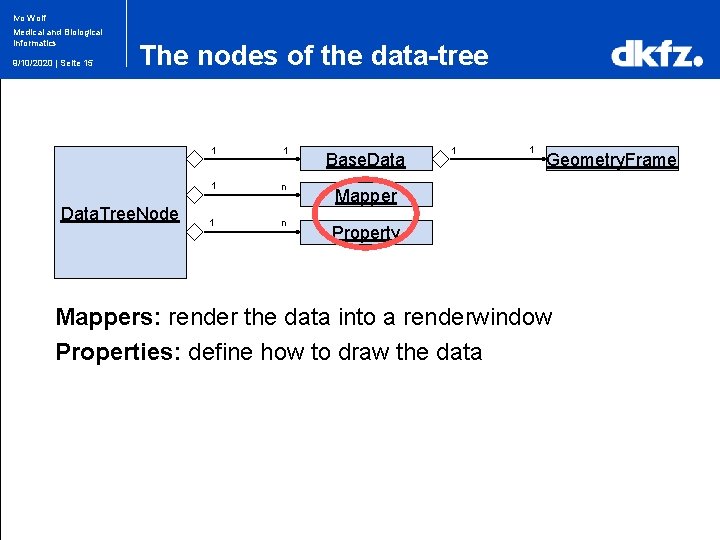
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 11 The nodes of the data-tree 1 1 Base. Data 1 1 Geometry. Frame Data. Tree. Node // creating an iterator to the data-tree mitk: : Data. Tree. Pre. Order. Iterator iterator. To. Tree(tree); // adding nodes to the data-tree iterator. To. Tree. Add(an. Image. Node); iterator. To. Tree. Add(a. Surface. Node); iterator. To. Tree. Add(another. Image. Node); iterator. To. Tree. Add(a. Plane. Node);

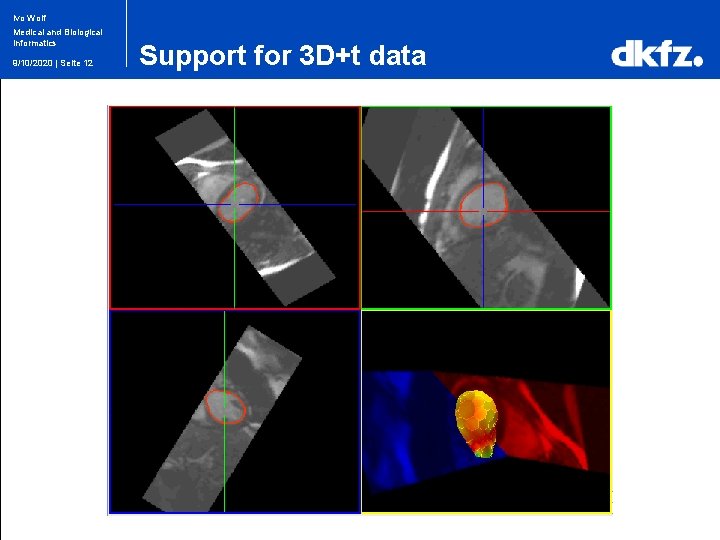
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 12 Support for 3 D+t data

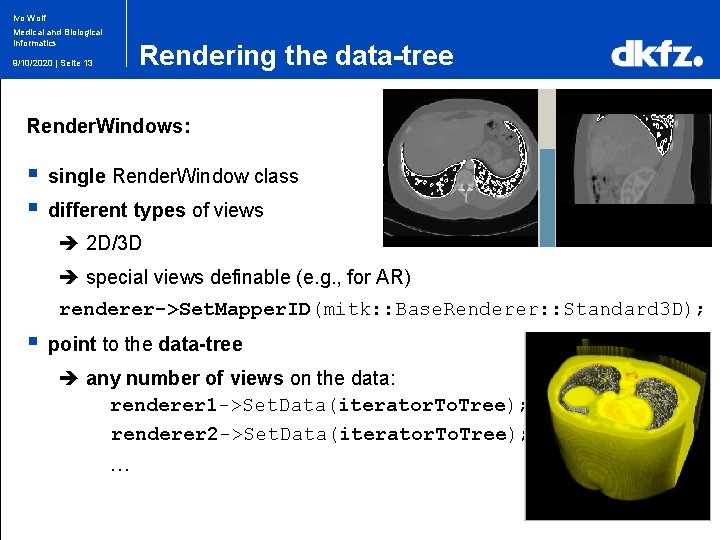
Ivo Wolf Medical and Biological Informatics Rendering the data-tree 9/10/2020 | Seite 13 Render. Windows: § § single Render. Window class different types of views 2 D/3 D special views definable (e. g. , for AR) renderer->Set. Mapper. ID(mitk: : Base. Renderer: : Standard 3 D); § point to the data-tree any number of views on the data: renderer 1 ->Set. Data(iterator. To. Tree); renderer 2 ->Set. Data(iterator. To. Tree); …

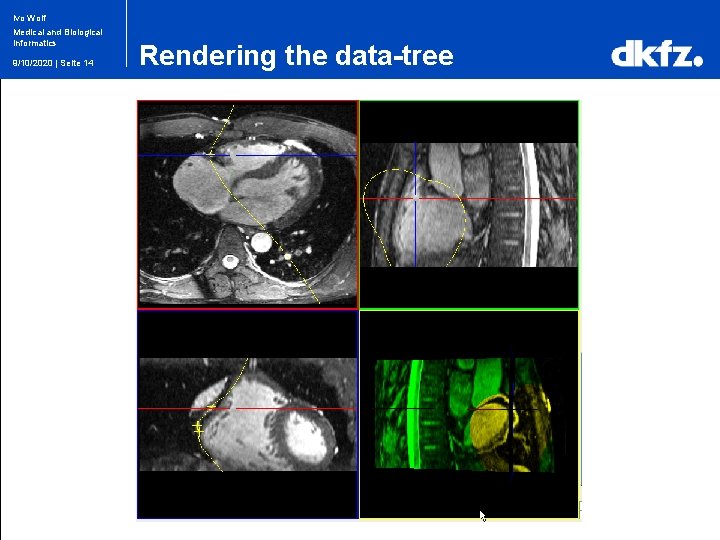
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 14 Rendering the data-tree

Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 15 The nodes of the data-tree Data. Tree. Node 1 1 Base. Data 1 n Mapper 1 n 1 1 Geometry. Frame Property Mappers: render the data into a renderwindow Properties: define how to draw the data

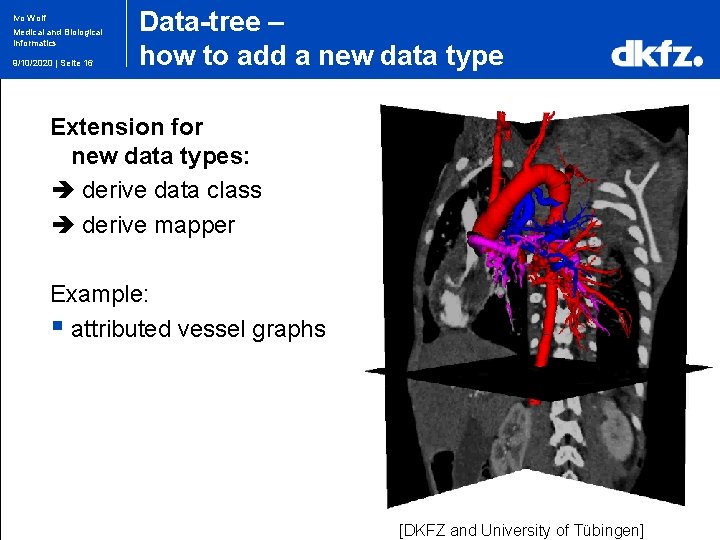
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 16 Data-tree – how to add a new data type Extension for new data types: derive data class derive mapper Example: § attributed vessel graphs [DKFZ and University of Tübingen]

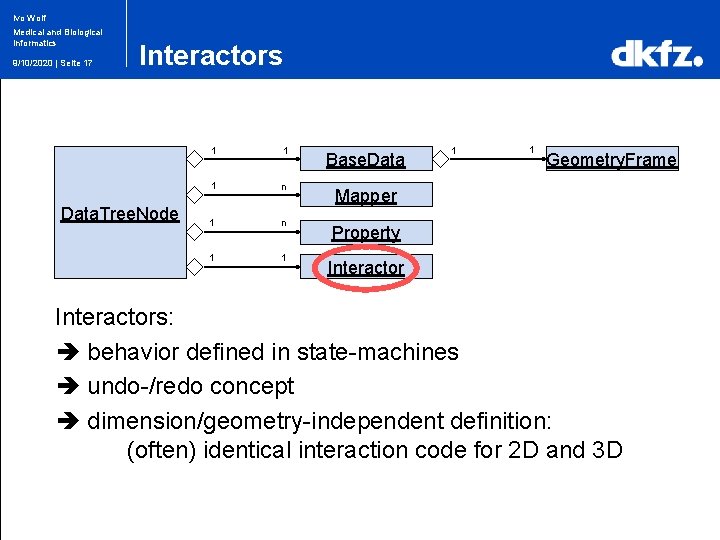
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 17 Interactors Data. Tree. Node 1 1 Base. Data 1 n Mapper 1 n 1 1 Geometry. Frame Property Interactors: behavior defined in state-machines undo-/redo concept dimension/geometry-independent definition: (often) identical interaction code for 2 D and 3 D

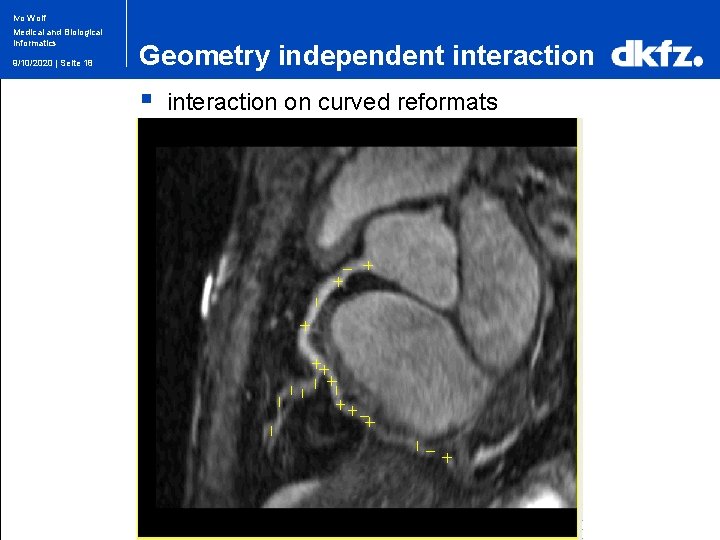
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 18 Geometry independent interaction § interaction on curved reformats

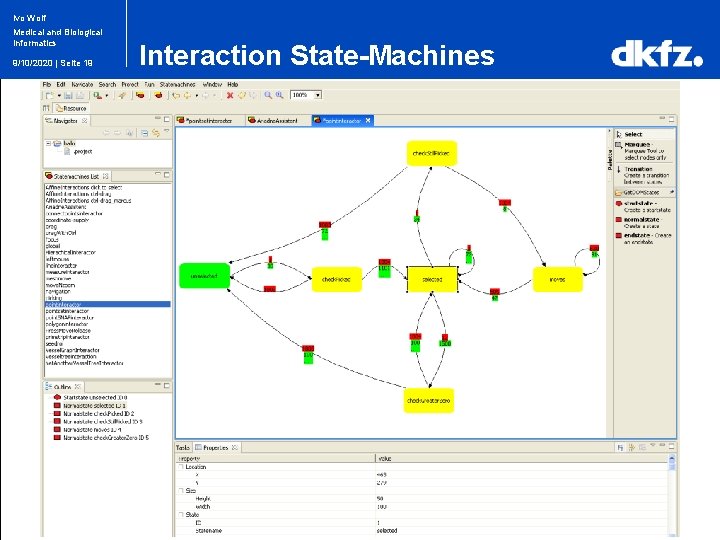
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 19 Interaction State-Machines

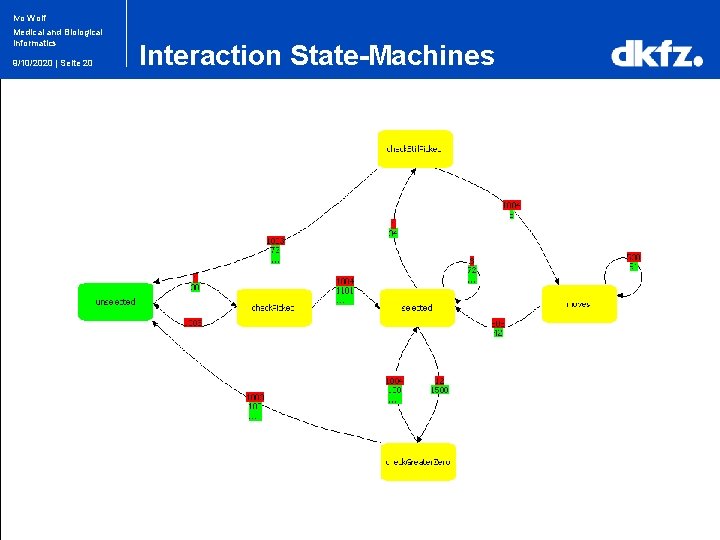
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 20 Interaction State-Machines

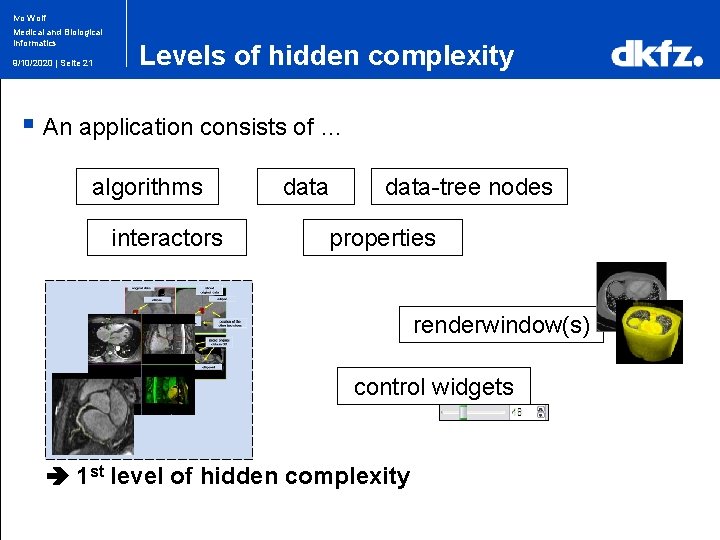
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 21 Levels of hidden complexity § An application consists of … algorithms interactors data-tree nodes properties renderwindow(s) control widgets 1 st level of hidden complexity

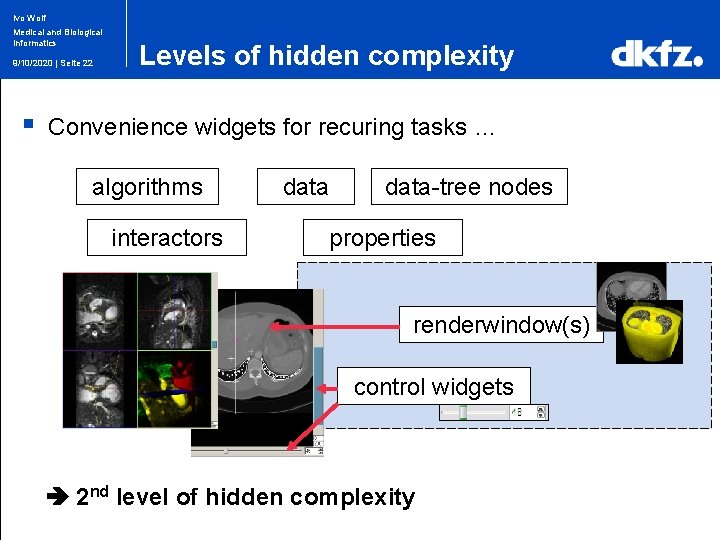
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 22 § Levels of hidden complexity Convenience widgets for recuring tasks … algorithms interactors data-tree nodes properties renderwindow(s) control widgets 2 nd level of hidden complexity

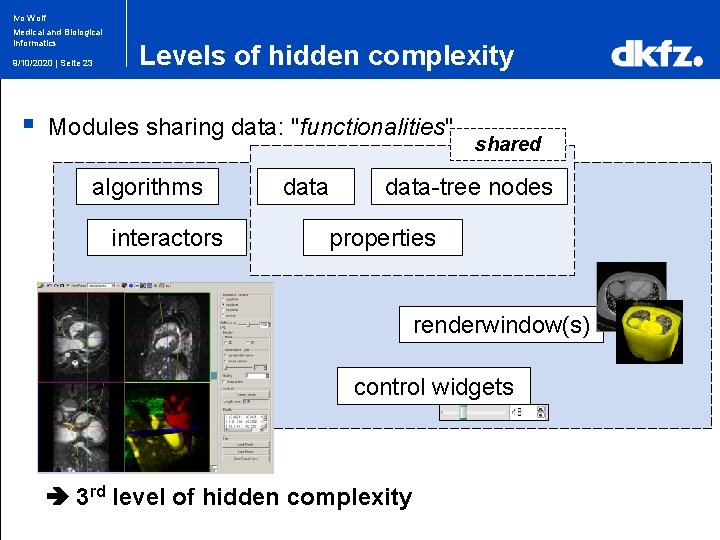
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 23 § Levels of hidden complexity Modules sharing data: "functionalities" algorithms interactors data shared data-tree nodes properties renderwindow(s) control widgets 3 rd level of hidden complexity

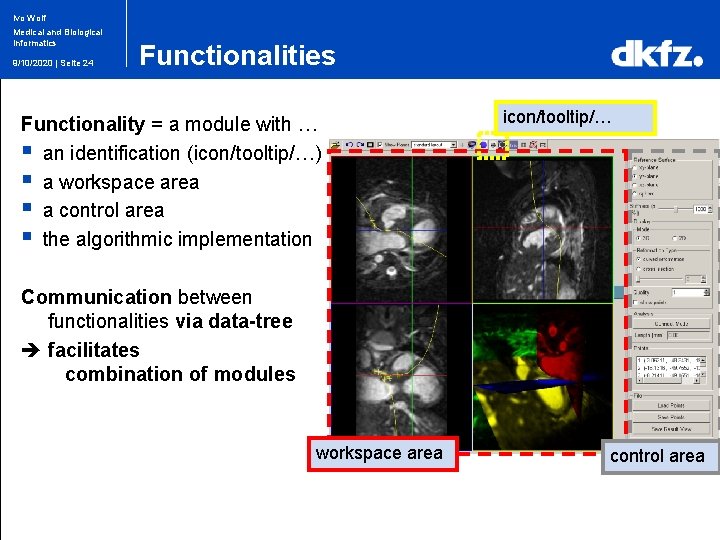
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 24 Functionalities Functionality = a module with … § an identification (icon/tooltip/…) § a workspace area § a control area § the algorithmic implementation icon/tooltip/… Communication between functionalities via data-tree facilitates combination of modules workspace area control area

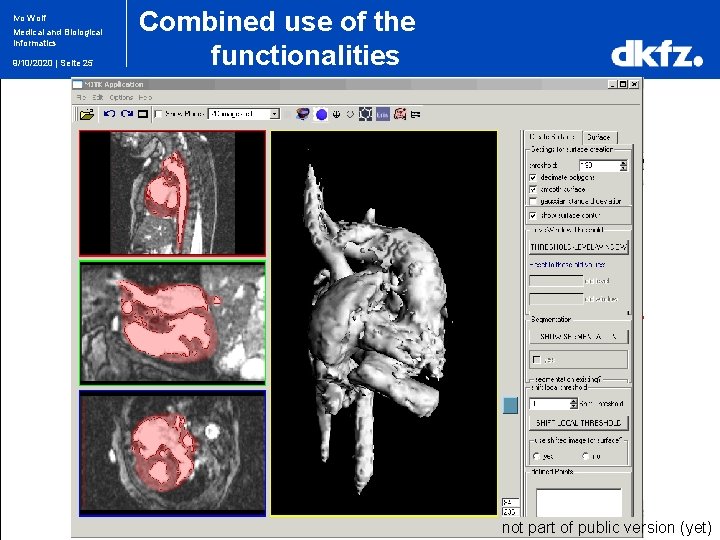
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 25 Combined use of the functionalities not part of public version (yet)

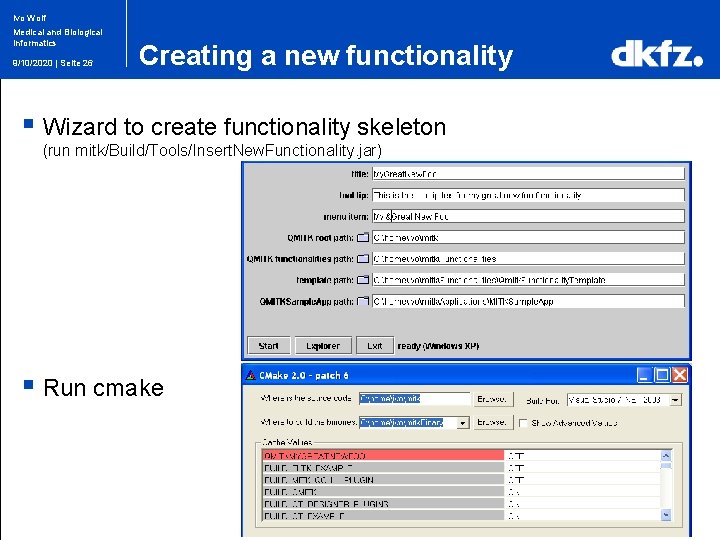
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 26 Creating a new functionality § Wizard to create functionality skeleton (run mitk/Build/Tools/Insert. New. Functionality. jar) § Run cmake

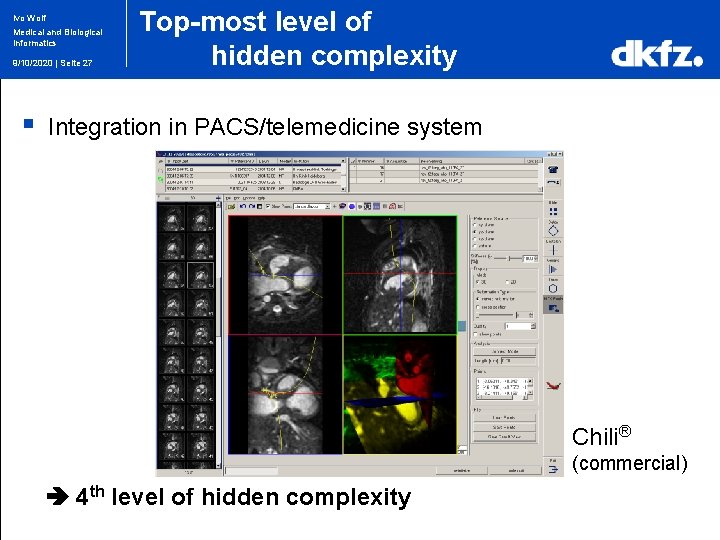
Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 27 § Top-most level of hidden complexity Integration in PACS/telemedicine system Chili® (commercial) 4 th level of hidden complexity

Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 28 www. mitk. org Download options: § anonymous cvs § zipped archive (v 0. 4 of Oct 21 st) Tutorial: § click on "Documentation"

Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 29 MITK – Summary § Supports development of interactive systems from the toolkit level Toolkit, not an application or development environment § Re-use of ITK/VTK code and concepts § Coordination of visualizations and interactions § Improved 2 D support (compared to basic VTK) § Adds more high-level interaction capabilities § Support for 3 D+t data § Different layers of hidden complexity § Facilitates re-use of high-level modules

Ivo Wolf Medical and Biological Informatics 9/10/2020 | Seite 30 Thank you Acknowledgement This work was supported in part within the grant SFB 414 "Computer- and sensor-supported surgery" of the DFG (German Research Foundation)