The MGED Ontology Is An Experimental Ontology BioOntologies

The MGED Ontology Is An Experimental Ontology Bio-Ontologies Aug 8, 2002 Chris Stoeckert, Helen Parkinson and the MGED Ontology Working Group
MGED Mission Statement • The Microarray Gene Expression Data (MGED) society is an international organization for facilitating the sharing of microarray data from functional genomics and proteomics experiments. • MGED was established as a grass roots movement in a meeting in November 1999 in Cambridge, UK • Current tasks involve establishing standards for microarray data annotation and representation, facilitating the creation of microarray databases and providing infrastructure for dissemination of experimental and data transformation protocols • Long term goals for the future will extend the mission to other functional genomics and proteomics high throughput technologies.

http: //www. mged. org

An Experimental Ontology • An ontology for microarray experiments – Not an ontology of life but of experiments – Parts are applicable to describing experiments in general • Our approach to interfacing with other ontologies is “experimental” – Not mapping terms from related ontologies – Provide a framework to hang other ontologies off of • Know where to find different types of annotation • How to interpret that annotation
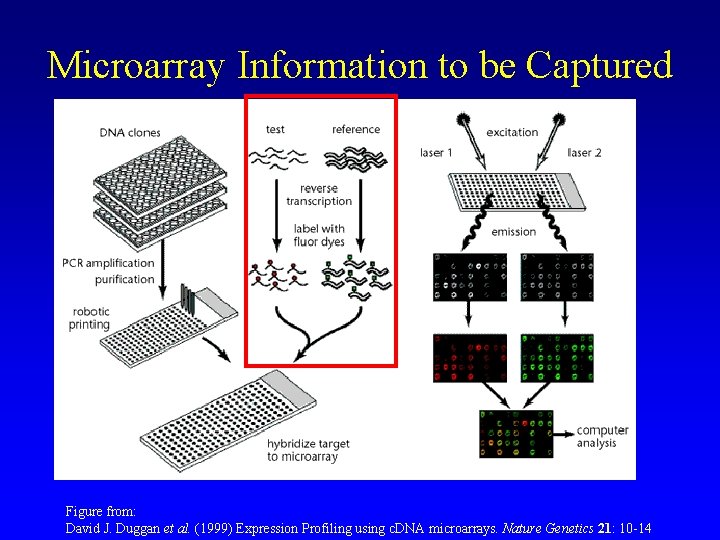
Microarray Information to be Captured Figure from: David J. Duggan et al. (1999) Expression Profiling using c. DNA microarrays. Nature Genetics 21: 10 -14
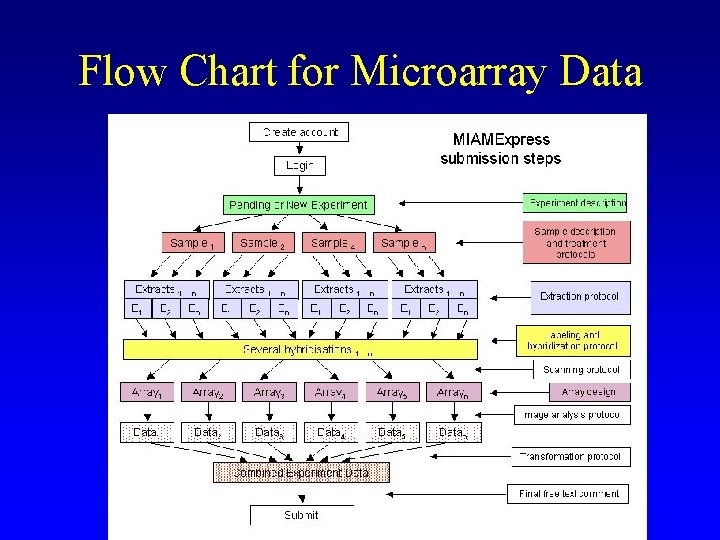
Flow Chart for Microarray Data

Minimal Information About a Microarray Experiment (MIAME) • Provides the concepts for the ontology • Array design description – Common features of the array as the whole, and the description of each array design elements (e. g. , each spot) • Gene expression experiment description – – Experimental design Samples used, extract preparation and labeling Hybridization procedures and parameters Measurement data and specifications of data processing • See Brazma et al Nature Genetics 2001 and http: //www. mged. org/Workgroups/MIAME/miame. html
MIAME Section on Samples (Biomaterials) • • Biosource properties – Organism – Contact details for sample – Descriptors relevant to the particular sample, such as • Sex • Age • Developmental stage • Organism part (tissue) • Cell type • Animal/ plant strain or line • Genetic variation (e. g. , gene knockout, transgenic variation) • Individual genetic characteristics (e. g. , disease alleles, polymorphisms) • Disease state or nornal • Is additional clinical information available (link) • The individual (for interrelation of the samples in the experiment) Biomaterial manipulations: laboratory protocol, including relevant parameters, e. g. , – Growth conditions – In vivo treatments (organism or individual treatments) – In vitro treatments (cell culture conditions) – Treatment type (e. g. , small molecule, heat shock, cold shock, food deprivation) – Compound – Separation technique (e. g. , none, trimming, microdissection, FACS)
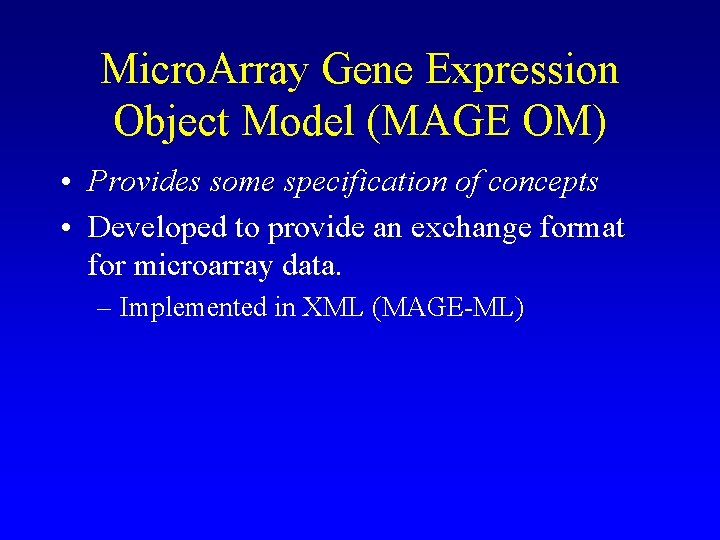
Micro. Array Gene Expression Object Model (MAGE OM) • Provides some specification of concepts • Developed to provide an exchange format for microarray data. – Implemented in XML (MAGE-ML)
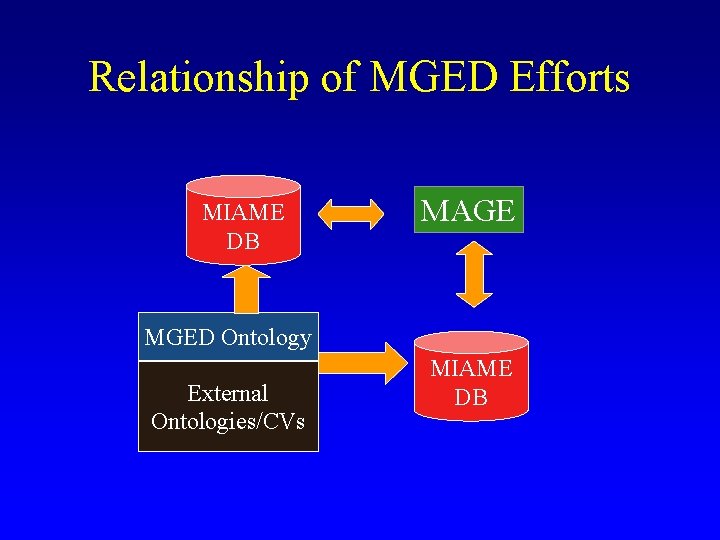
Relationship of MGED Efforts MIAME DB MAGE MGED Ontology External Ontologies/CVs MIAME DB

The MGED Ontology Working Group • Acts through – a mailing list of over 250 – working group meetings organized at conferences like ISMB and of course MGED • Collects resources (dictionaries, controlled vocabularies, ontologies) for terms to describe microarray experiments – Sample (biomaterial) – Experimental conditions (treatments) – Experimental design (study design)
The MGED Ontology Home Page http: //www. cbil. upenn. edu/Ontology
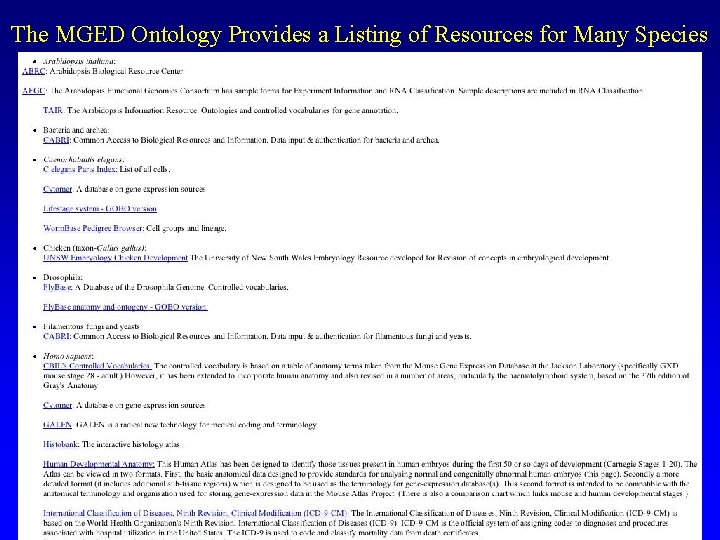
The MGED Ontology Provides a Listing of Resources for Many Species
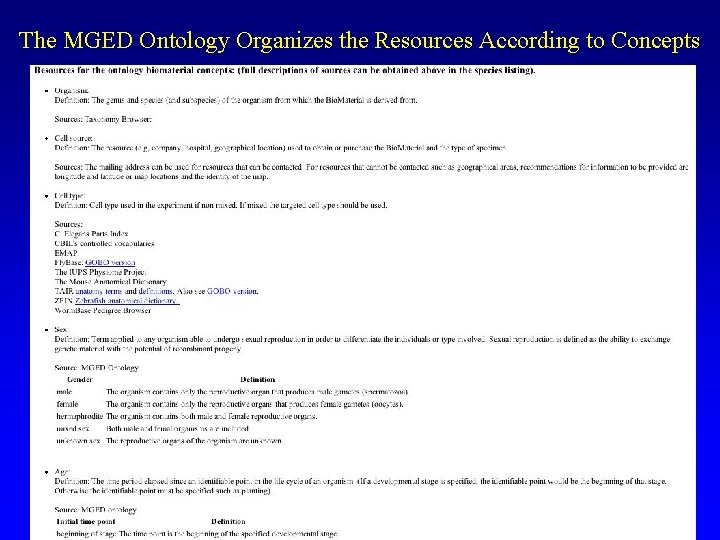
The MGED Ontology Organizes the Resources According to Concepts
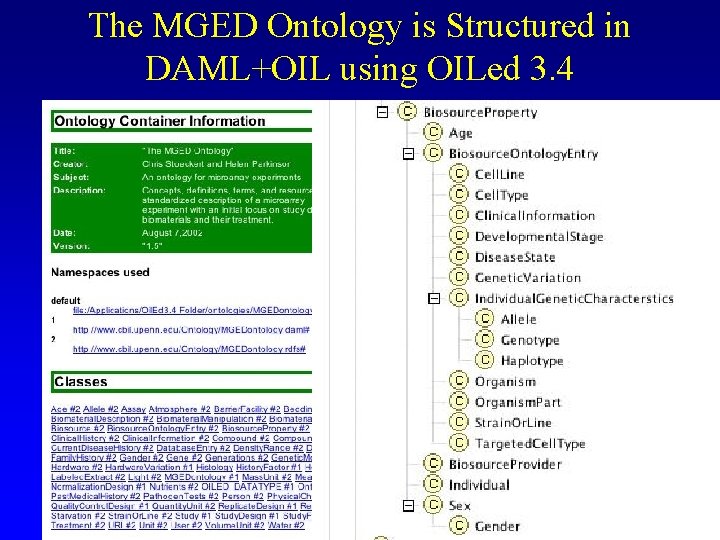
The MGED Ontology is Structured in DAML+OIL using OILed 3. 4
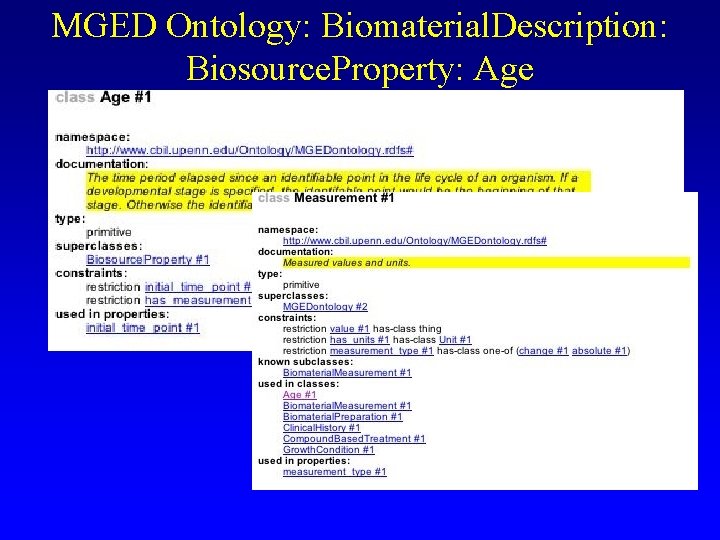
MGED Ontology: Biomaterial. Description: Biosource. Property: Age
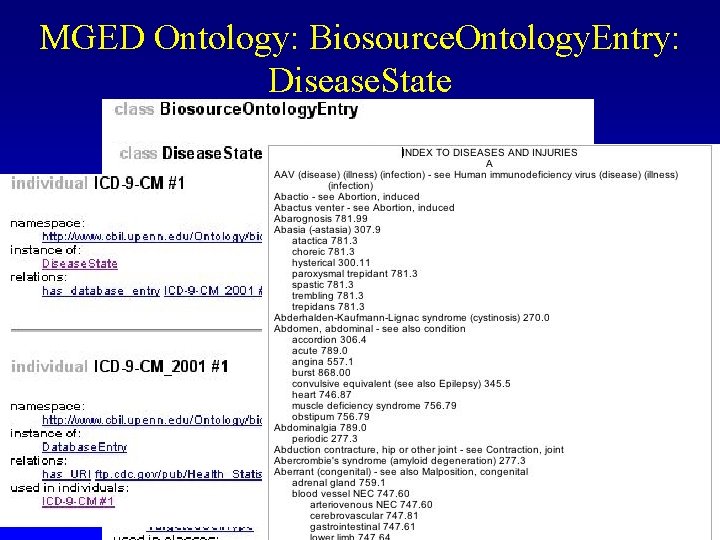
MGED Ontology: Biosource. Ontology. Entry: Disease. State
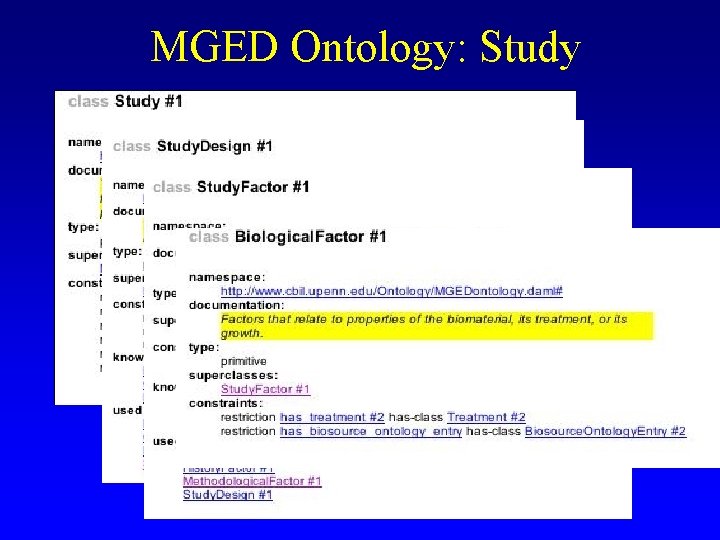
MGED Ontology: Study

MGED Ontology Use Cases • Make it easier and more accurate to annotate a microarray experiment. – Build forms that provide menus of terms and links to external resources. See MIAMEexpress! – Only ask for relevant terms and fill in terms that can be inferred. • Use structured fields and controlled terms to query databases. – Return a summary of all experiments that use a specified type of biosource. – Return a summary of all experiments done examining effects of a specified treatment • ? Aid in experiment design by providing parameters to consider about samples, organization of treatments. • ? Use to check if “MIAME-compliant. ” – Assess only fields that are relevant – Check for proper use of terms • ? Build gene networks based on biomaterial description – Use structured descriptions to cluster, build models, etc.
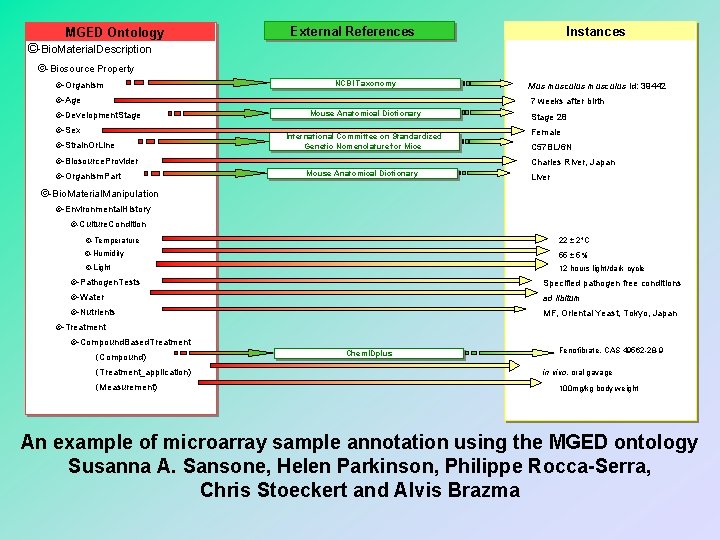
MGED Ontology External References Instances ©-Bio. Material. Description ©-Biosource Property ©-Organism NCBI Taxonomy ©-Age Mus musculus id: 39442 7 weeks after birth ©-Development. Stage ©-Sex ©-Strain. Or. Line Mouse Anatomical Dictionary International Committee on Standardized Genetic Nomenclature for Mice ©-Biosource. Provider ©-Organism. Part Stage 28 Female C 57 BL/6 N Charles River, Japan Mouse Anatomical Dictionary Liver ©-Bio. Material. Manipulation ©-Environmental. History ©-Culture. Condition ©-Temperature 22 2 C ©-Humidity 55 5% ©-Light 12 hours light/dark cycle ©-Pathogen. Tests Specified pathogen free conditions ©-Water ad libitum ©-Nutrients MF, Oriental Yeast, Tokyo, Japan ©-Treatment ©-Compound. Based. Treatment (Compound) (Treatment_application) (Measurement) Chem. IDplus Fenofibrate, CAS 49562 -28 -9 in vivo, oral gavage 100 mg/kg body weight An example of microarray sample annotation using the MGED ontology Susanna A. Sansone, Helen Parkinson, Philippe Rocca-Serra, Chris Stoeckert and Alvis Brazma

The MGED Ontology in Action: MIAMExpress
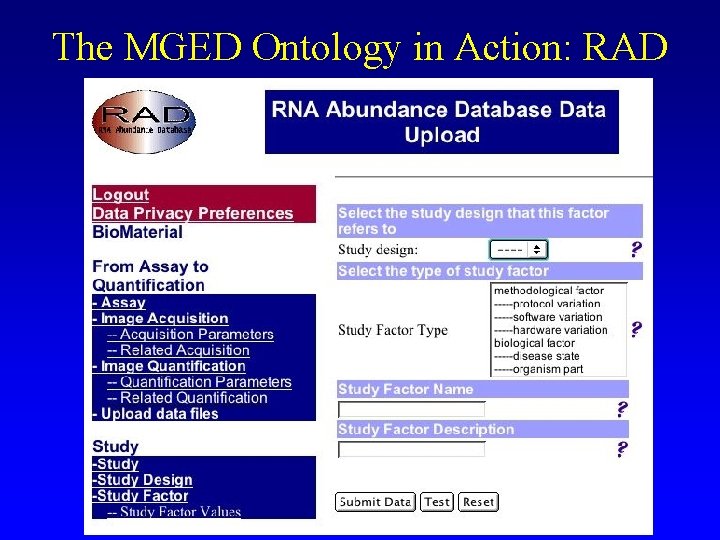
The MGED Ontology in Action: RAD

Summary • The MGED Ontology is being developed within the microarray community to provide consistent terminology for experiments. • This community effort has resulted in a list of multiple resources for many species. • The list is organized by defined concepts and augmented with terms for widely applicable concepts (e. g. , “age”, “sex”). • The concepts are structured in DAML+OIL and available in other formats (rdfs) • The MGED Ontology is a work in progress – More instances (create IDs) – Constraints – Concepts for other parts of microarray experiment

http: //www. ebi. ac. uk/SOFG
- Slides: 26