The genome functions in the context of chromatin
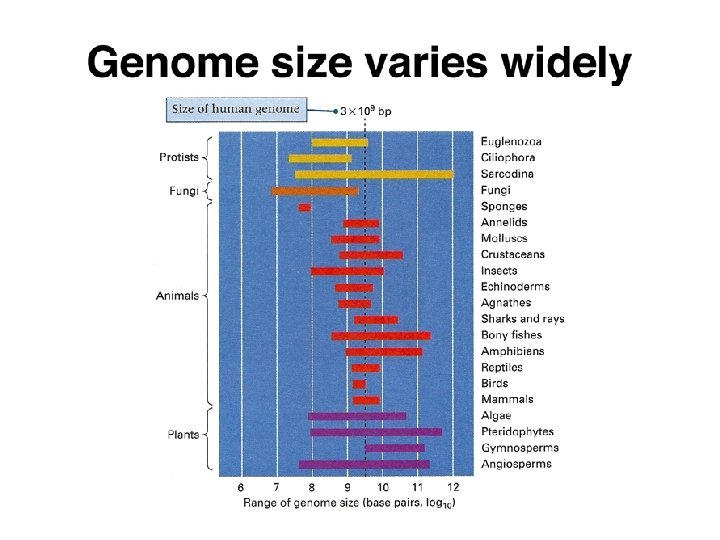
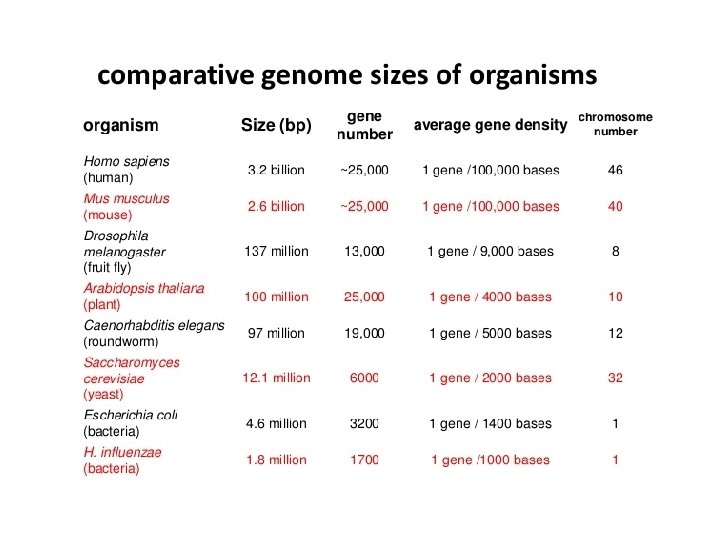







- Slides: 45

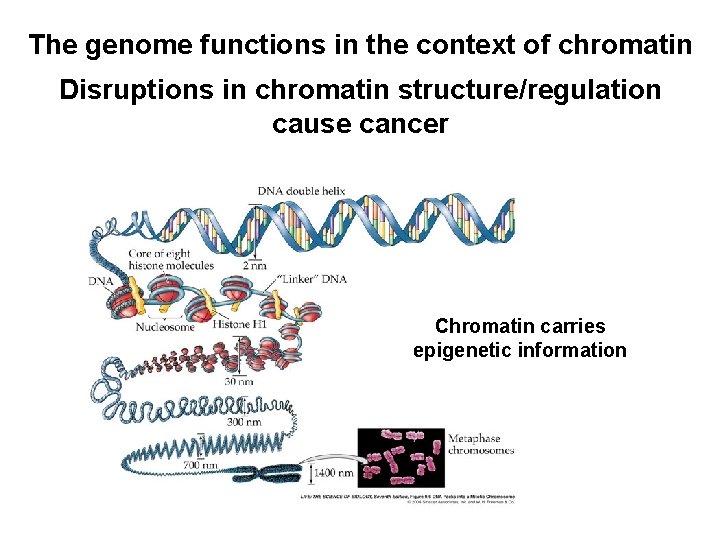
The genome functions in the context of chromatin Disruptions in chromatin structure/regulation cause cancer Chromatin carries epigenetic information




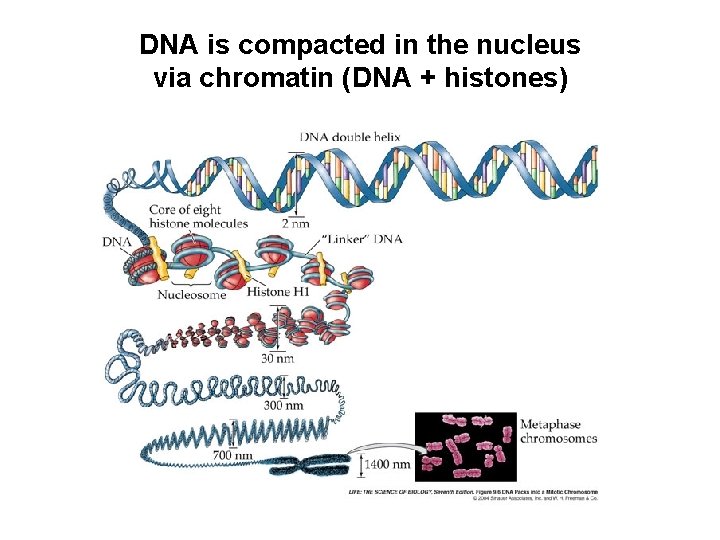
DNA is compacted in the nucleus via chromatin (DNA + histones)

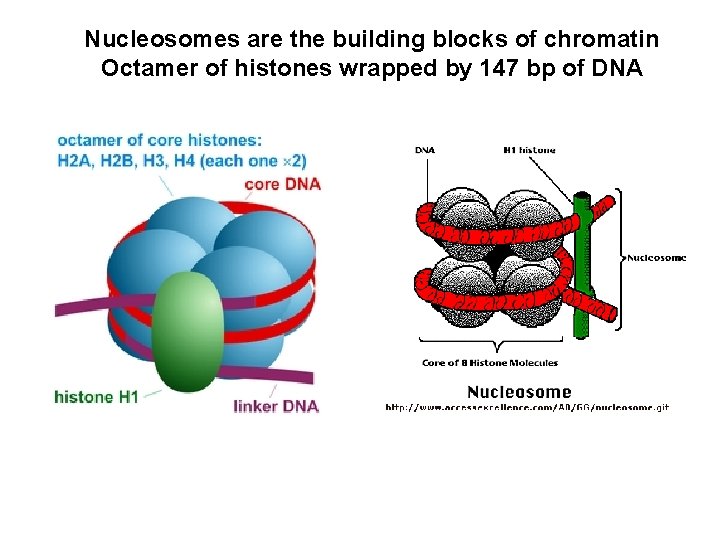
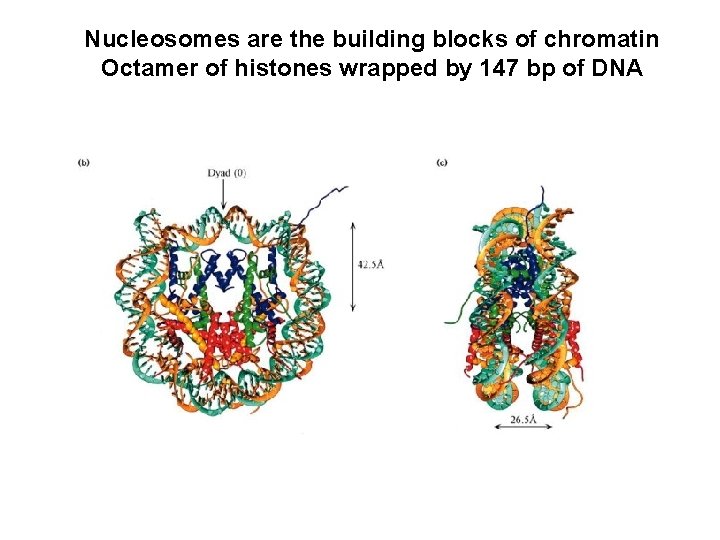
Nucleosomes are the building blocks of chromatin Octamer of histones wrapped by 147 bp of DNA

Nucleosomes are the building blocks of chromatin Octamer of histones wrapped by 147 bp of DNA

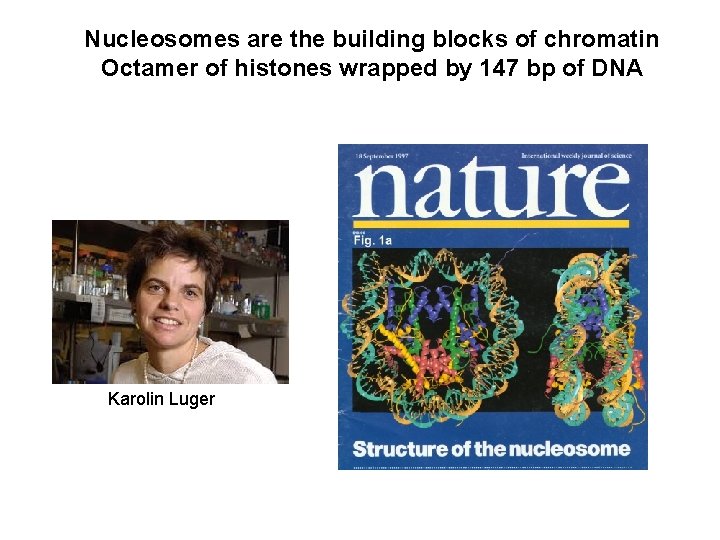
Nucleosomes are the building blocks of chromatin Octamer of histones wrapped by 147 bp of DNA Karolin Luger

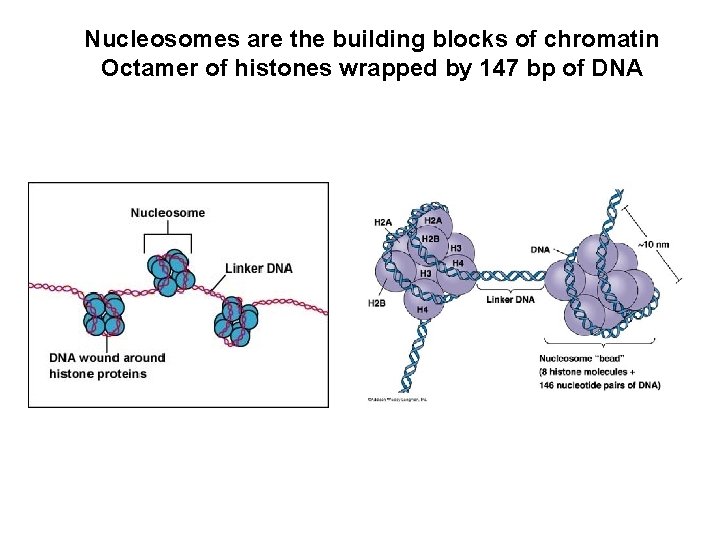
Nucleosomes are the building blocks of chromatin Octamer of histones wrapped by 147 bp of DNA

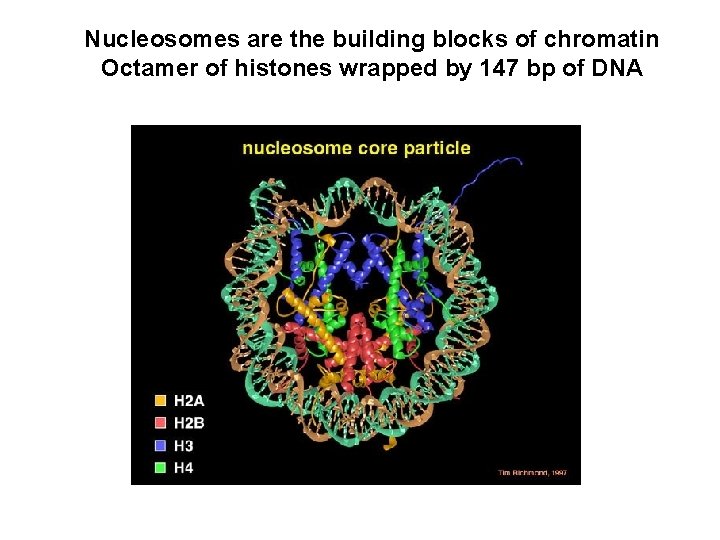
Nucleosomes are the building blocks of chromatin Octamer of histones wrapped by 147 bp of DNA

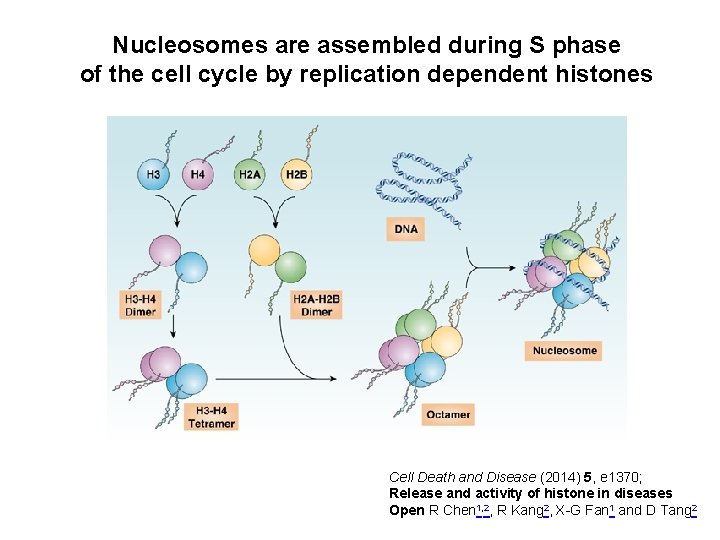
Nucleosomes are assembled during S phase of the cell cycle by replication dependent histones Cell Death and Disease (2014) 5, e 1370; Release and activity of histone in diseases Open R Chen 1, 2, R Kang 2, X-G Fan 1 and D Tang 2

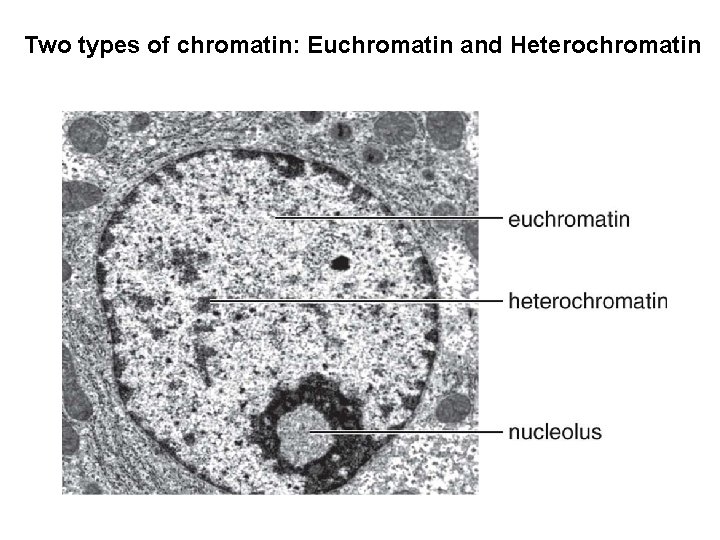
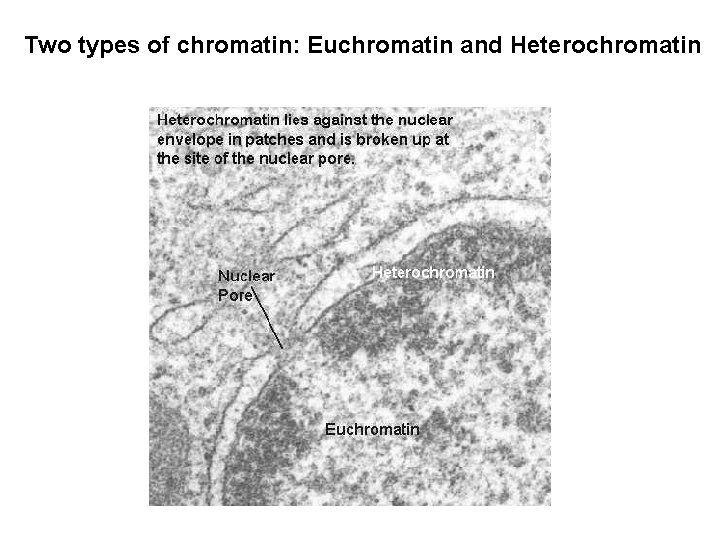
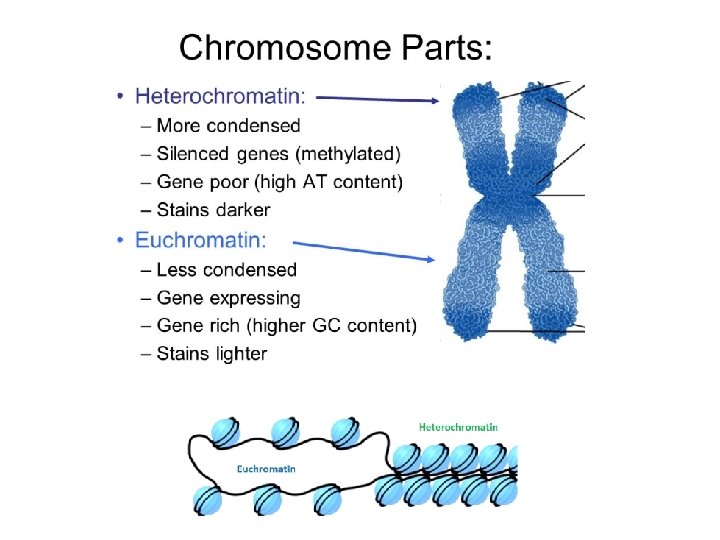
Two types of chromatin: Euchromatin and Heterochromatin

Two types of chromatin: Euchromatin and Heterochromatin


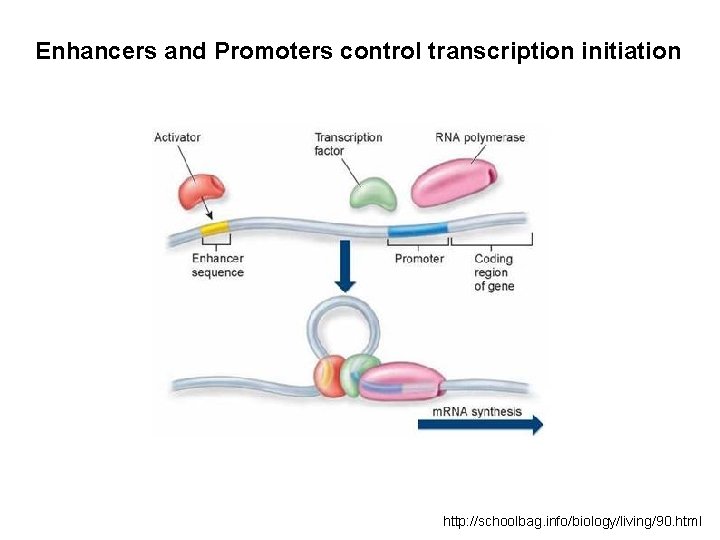
Enhancers and Promoters control transcription initiation http: //schoolbag. info/biology/living/90. html

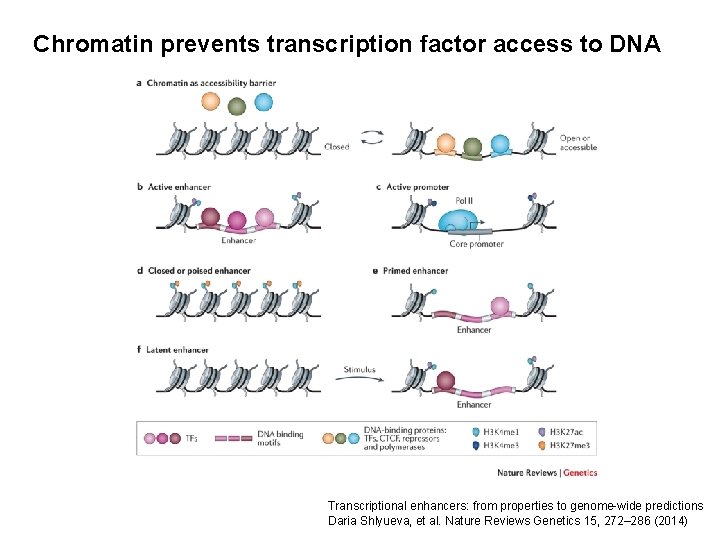
Chromatin prevents transcription factor access to DNA Transcriptional enhancers: from properties to genome-wide predictions Daria Shlyueva, et al. Nature Reviews Genetics 15, 272– 286 (2014)

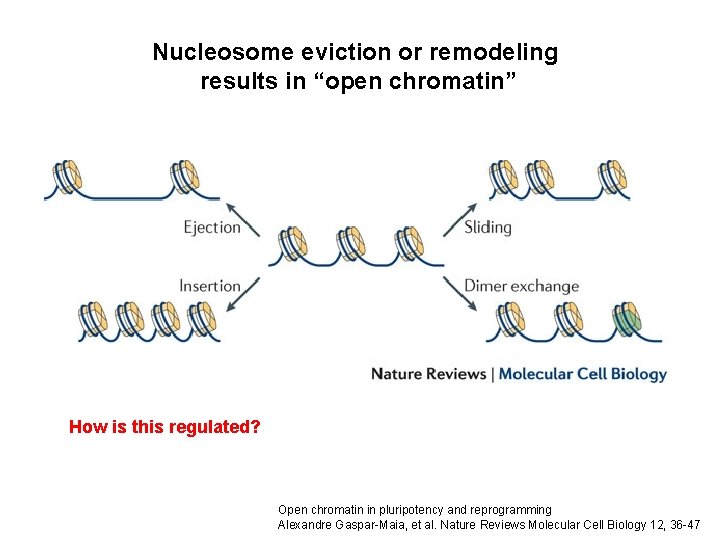
Nucleosome eviction or remodeling results in “open chromatin” How is this regulated? Open chromatin in pluripotency and reprogramming Alexandre Gaspar-Maia, et al. Nature Reviews Molecular Cell Biology 12, 36 -47

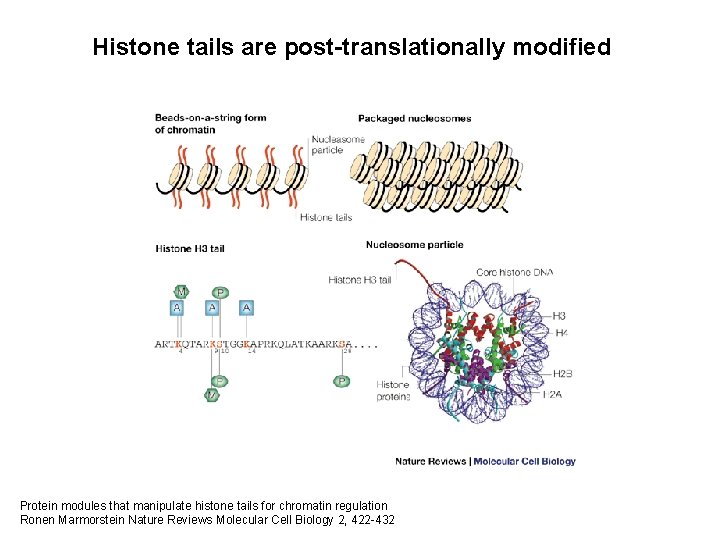
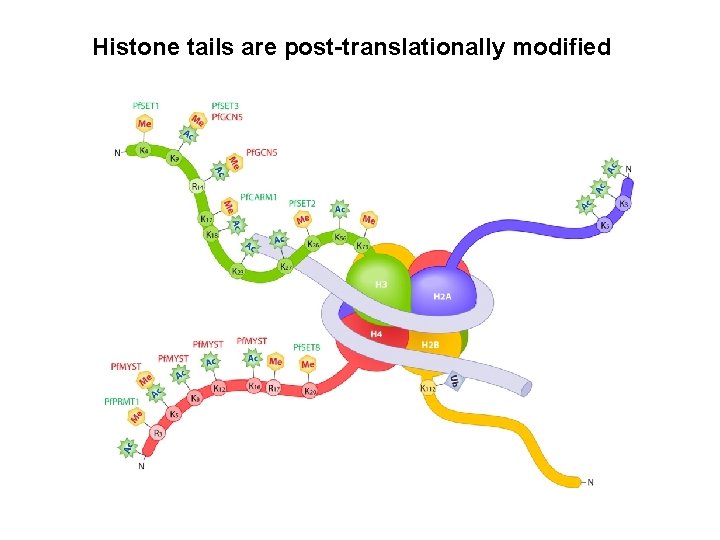
Histone tails are post-translationally modified Protein modules that manipulate histone tails for chromatin regulation Ronen Marmorstein Nature Reviews Molecular Cell Biology 2, 422 -432

Histone tails are post-translationally modified

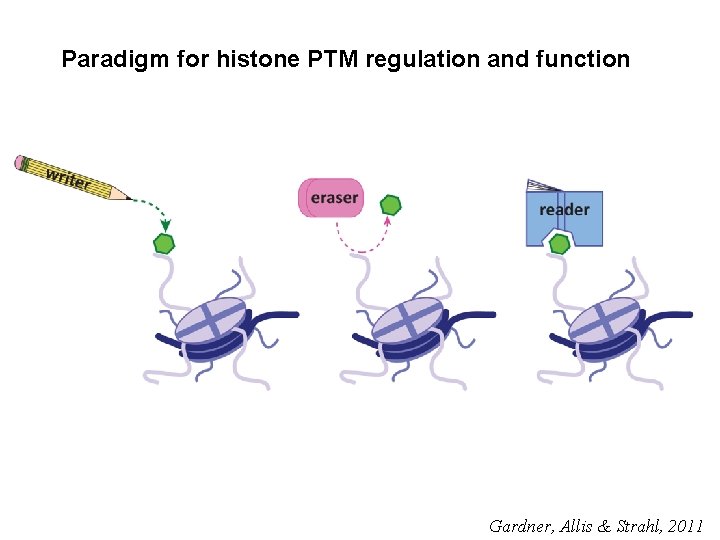
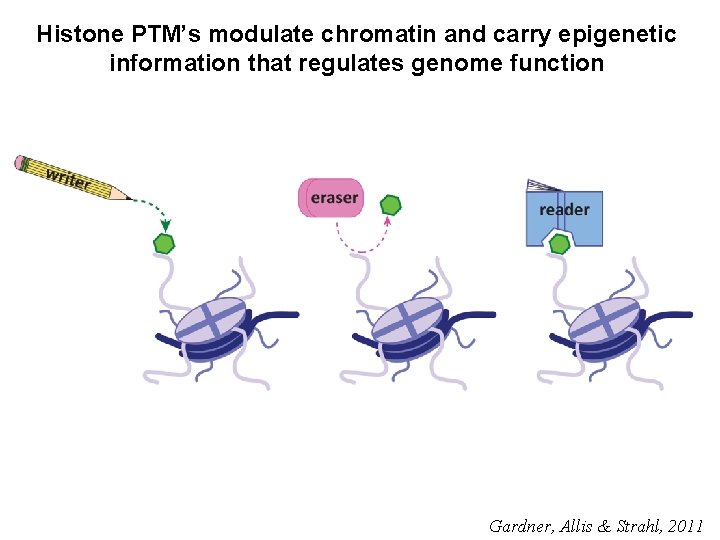
Paradigm for histone PTM regulation and function Gardner, Allis & Strahl, 2011

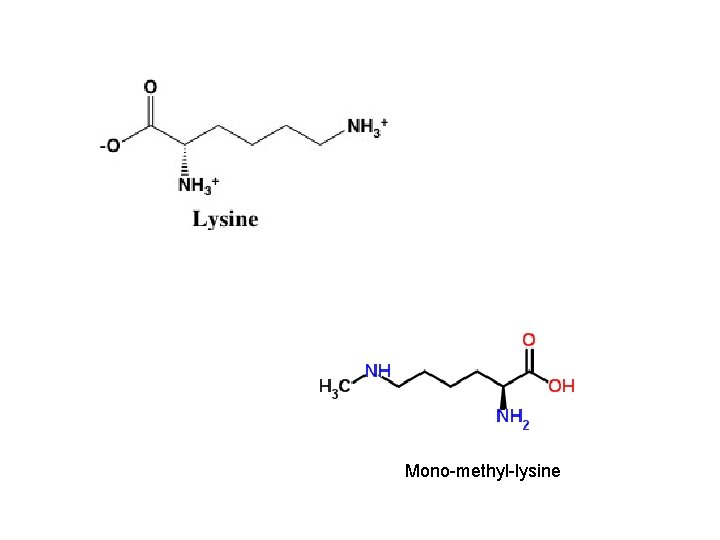
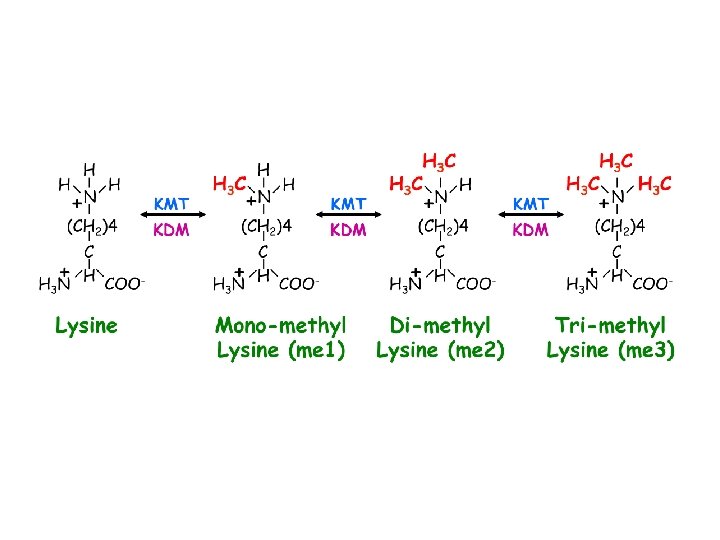
Mono-methyl-lysine


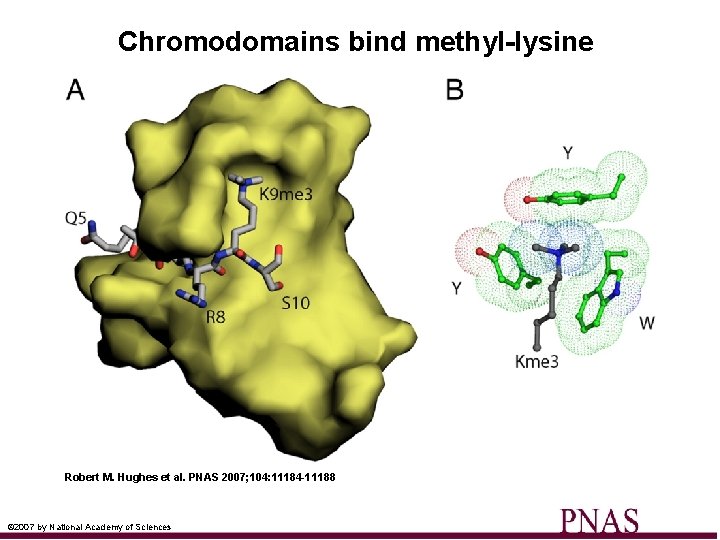
Chromodomains bind methyl-lysine Robert M. Hughes et al. PNAS 2007; 104: 11184 -11188 © 2007 by National Academy of Sciences

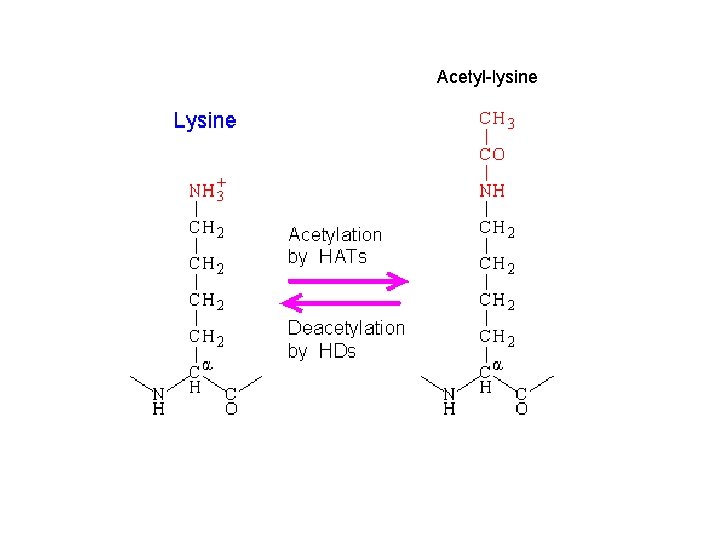
Acetyl-lysine

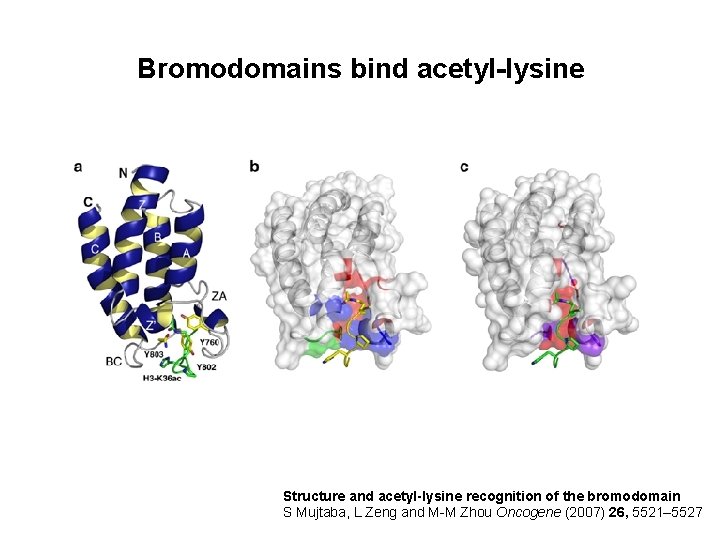
Bromodomains bind acetyl-lysine Structure and acetyl-lysine recognition of the bromodomain S Mujtaba, L Zeng and M-M Zhou Oncogene (2007) 26, 5521– 5527

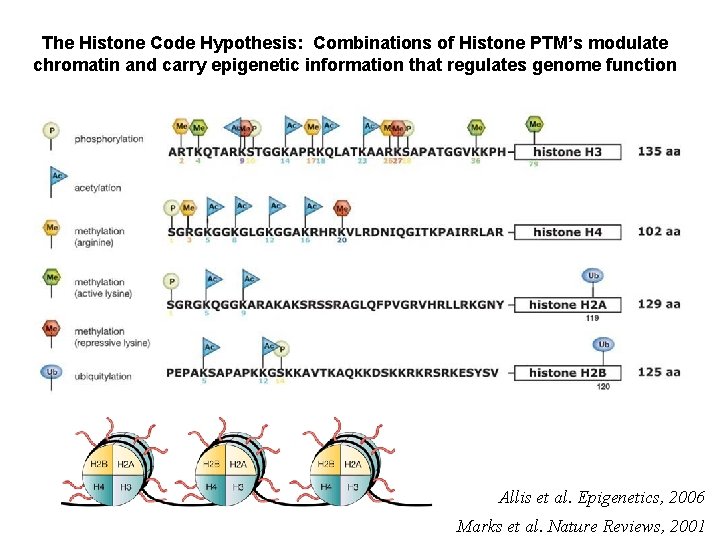
The Histone Code Hypothesis: Combinations of Histone PTM’s modulate chromatin and carry epigenetic information that regulates genome function Allis et al. Epigenetics, 2006 Marks et al. Nature Reviews, 2001

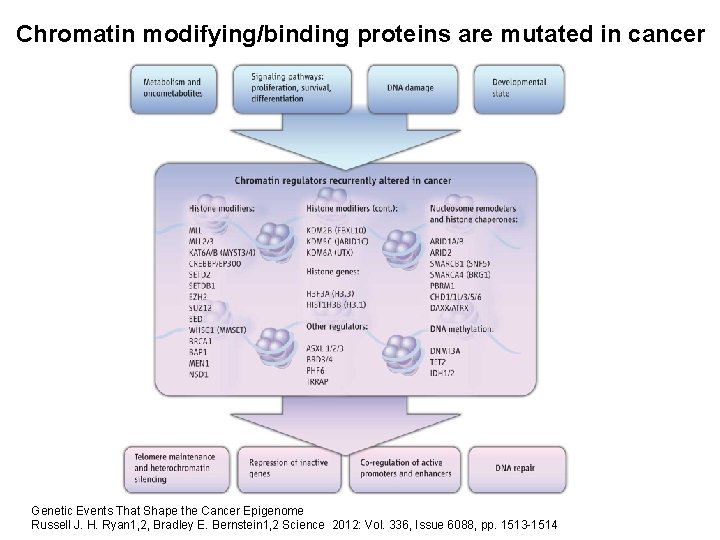
Chromatin modifying/binding proteins are mutated in cancer Genetic Events That Shape the Cancer Epigenome Russell J. H. Ryan 1, 2, Bradley E. Bernstein 1, 2 Science 2012: Vol. 336, Issue 6088, pp. 1513 -1514

Genetically testing the histone code hypothesis in an animal Greg Matera Dan Mc. Kay Brian Strahl Bill Marzluff The UNC Histone Replacement Group Departments of Biology, Genetics and Biochemistry & Biophysics Integrative Program for Biological and Genome Sciences Lineberger Comprehensive Cancer Center University of North Carolina at Chapel Hill

Histone PTM’s modulate chromatin and carry epigenetic information that regulates genome function Gardner, Allis & Strahl, 2011

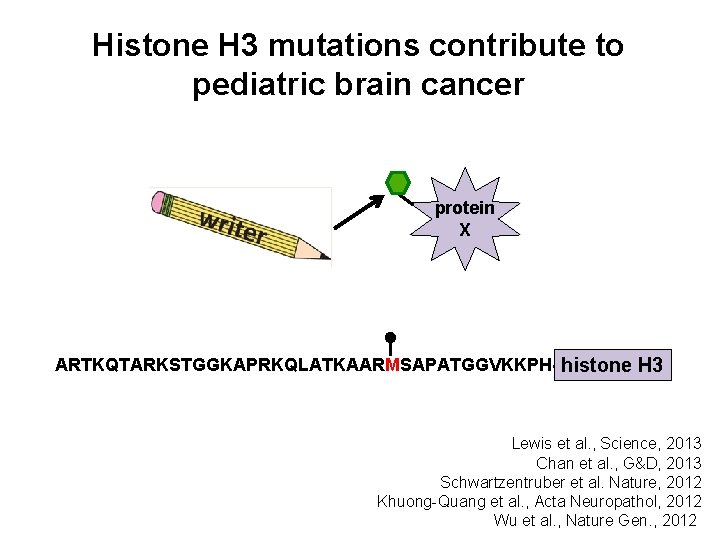
Writers can have non-histone substrates protein X ARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPH- histone H 3 Huang and Berger, 2008 Sims and Reinberg, 2008

Writers can have non-histone substrates So what causes the phenotype? ? ? protein X ARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPH- histone H 3

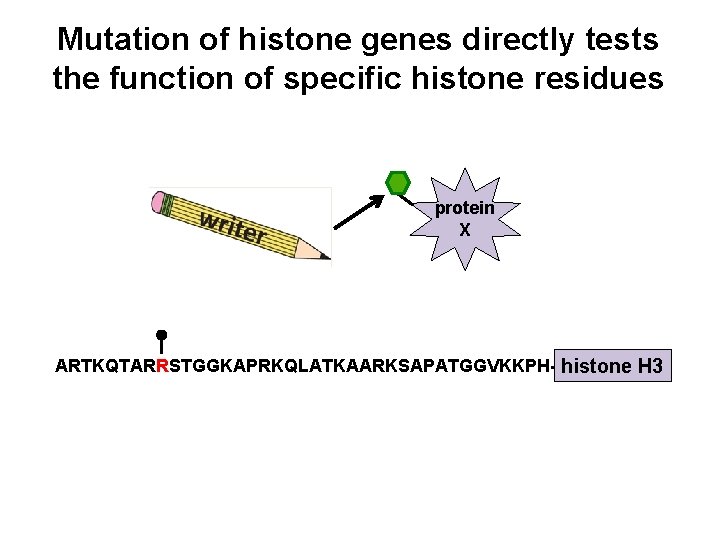
Mutation of histone genes directly tests the function of specific histone residues protein X ARTKQTARRSTGGKAPRKQLATKAARKSAPATGGVKKPH- histone H 3

Histone H 3 mutations contribute to pediatric brain cancer protein X ARTKQTARKSTGGKAPRKQLATKAARMSAPATGGVKKPH- histone H 3 Lewis et al. , Science, 2013 Chan et al. , G&D, 2013 Schwartzentruber et al. Nature, 2012 Khuong-Quang et al. , Acta Neuropathol, 2012 Wu et al. , Nature Gen. , 2012

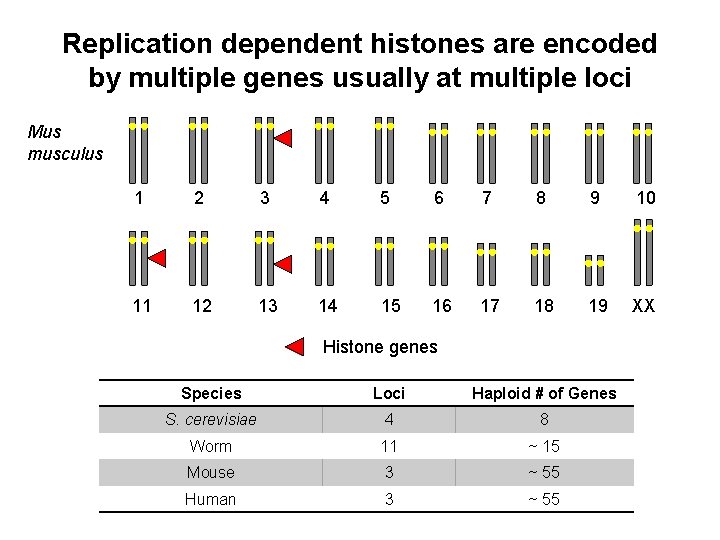
Replication dependent histones are encoded by multiple genes usually at multiple loci Mus musculus 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 XX Histone genes Species Loci Haploid # of Genes S. cerevisiae 4 8 Worm 11 ~ 15 Mouse 3 ~ 55 Human 3 ~ 55

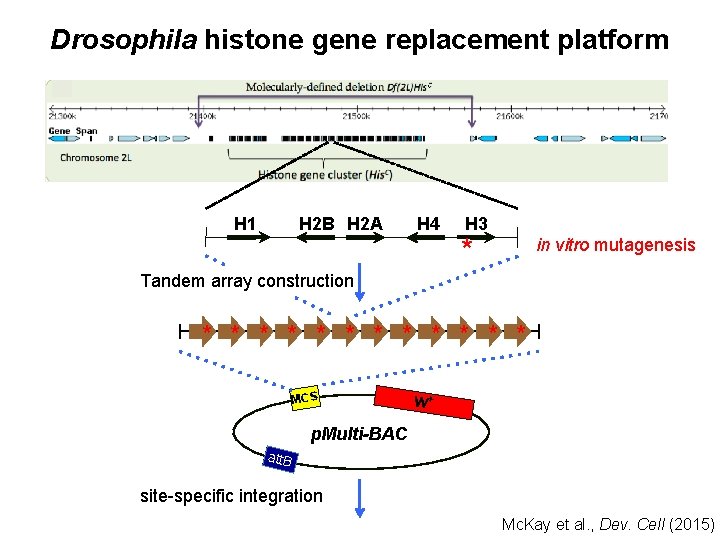
Drosophila histone gene replacement platform H 1 H 2 B H 2 A H 4 H 3 * in vitro mutagenesis Tandem array construction * * * MCS w+ p. Multi-BAC att. B site-specific integration Mc. Kay et al. , Dev. Cell (2015)

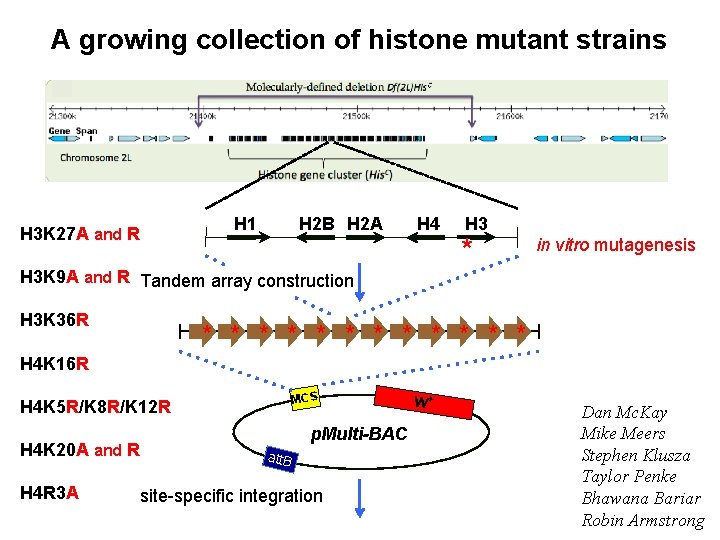
A growing collection of histone mutant strains H 1 H 3 K 27 A and R H 2 B H 2 A H 4 H 3 K 9 A and R Tandem array construction H 3 K 36 R H 3 * in vitro mutagenesis * * * H 4 K 16 R MCS H 4 K 5 R/K 8 R/K 12 R H 4 K 20 A and R H 4 R 3 A p. Multi-BAC att. B site-specific integration w+ Dan Mc. Kay Mike Meers Stephen Klusza Taylor Penke Bhawana Bariar Robin Armstrong

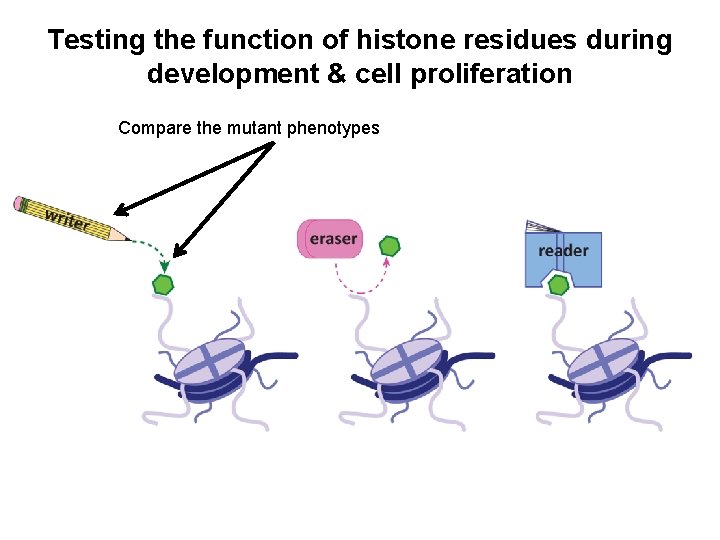
Testing the function of histone residues during development & cell proliferation Compare the mutant phenotypes

The role of H 4 K 20 in DNA replication and development Stephen Klusza (former LCCC fellow) The UNC Histone Replacement Group Departments of Biology, Genetics and Biochemistry & Biophysics Integrative Program for Biological and Genome Sciences Lineberger Comprehensive Cancer Center University of North Carolina at Chapel Hill

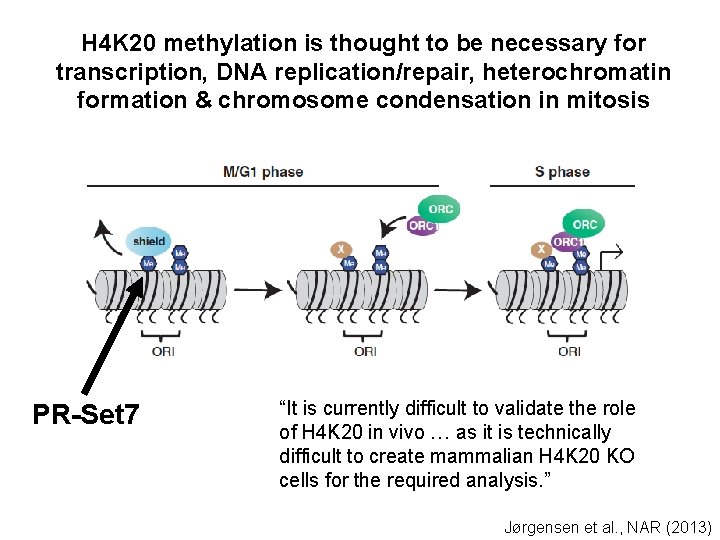
H 4 K 20 methylation is thought to be necessary for transcription, DNA replication/repair, heterochromatin formation & chromosome condensation in mitosis PR-Set 7 “It is currently difficult to validate the role of H 4 K 20 in vivo … as it is technically difficult to create mammalian H 4 K 20 KO cells for the required analysis. ” Jørgensen et al. , NAR (2013)

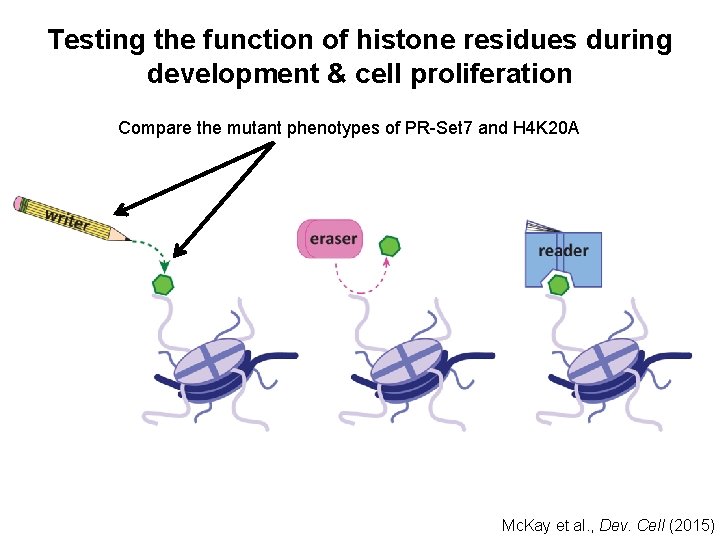
Testing the function of histone residues during development & cell proliferation Compare the mutant phenotypes of PR-Set 7 and H 4 K 20 A Mc. Kay et al. , Dev. Cell (2015)

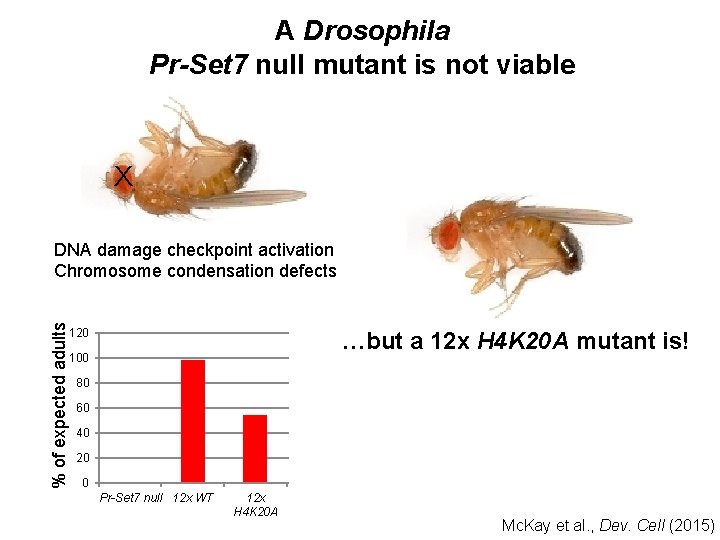
A Drosophila Pr-Set 7 null mutant is not viable X DNA damage checkpoint activation Chromosome condensation defects Sakaguchi et al. , JCB (2007)

A Drosophila Pr-Set 7 null mutant is not viable X % of expected adults DNA damage checkpoint activation Chromosome condensation defects 120 …but a 12 x H 4 K 20 A mutant is! 100 80 60 40 20 0 Pr-Set 7 null 12 x WT 12 x H 4 K 20 A Pr-Set 7 hypo Mc. Kay et al. , Dev. Cell (2015)

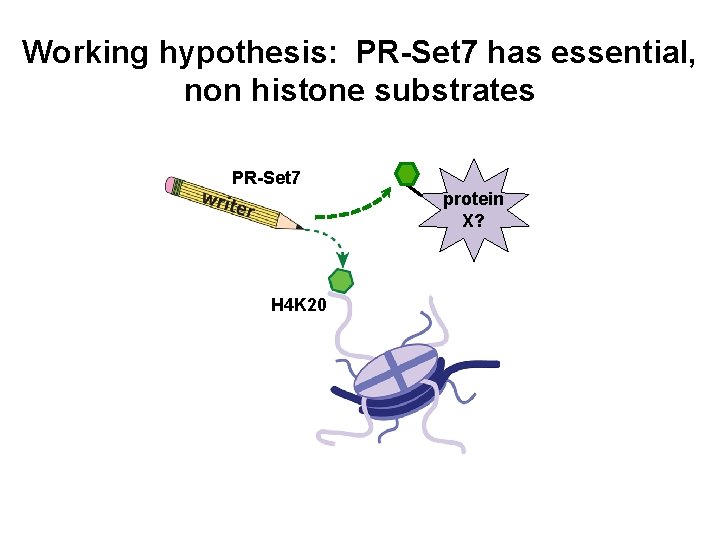
Working hypothesis: PR-Set 7 has essential, non histone substrates PR-Set 7 protein X? H 4 K 20

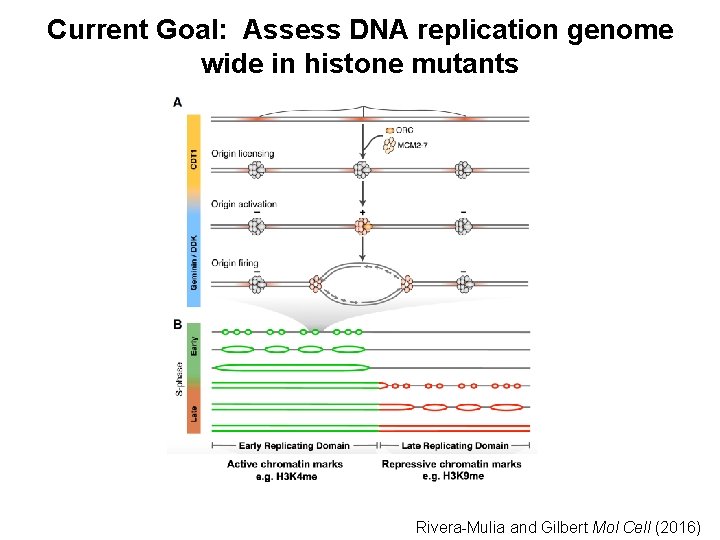
Current Goal: Assess DNA replication genome wide in histone mutants Rivera-Mulia and Gilbert Mol Cell (2016)

It takes a village… Matera Lab Duronio Lab Bhawana Bariar Mike Meers Taylor Penke Special Thanks: Robin Armstrong Stephen Klusza Alf Herzig Bill Marzluff Mc. Kay Lab Raquel Oliveira Spencer Nystrom Ruth Steward Mary Leatham-Jensen Chris Uyehara Strahl Lab Stephen Mc. Daniel Raghuvar Dronamraiu Funding: LCCC, NIH Katie Stallard-Bolling