The genetic code Nucleic acids Correspondence the genetic

- Slides: 20

The genetic code Nucleic acids Correspondence = the genetic code Amino acids Codon = triplet of three bases which encodes an amino acid 64 possible codons = 43 each of 4 nucleotides can occupy each of 3 positions in the codon

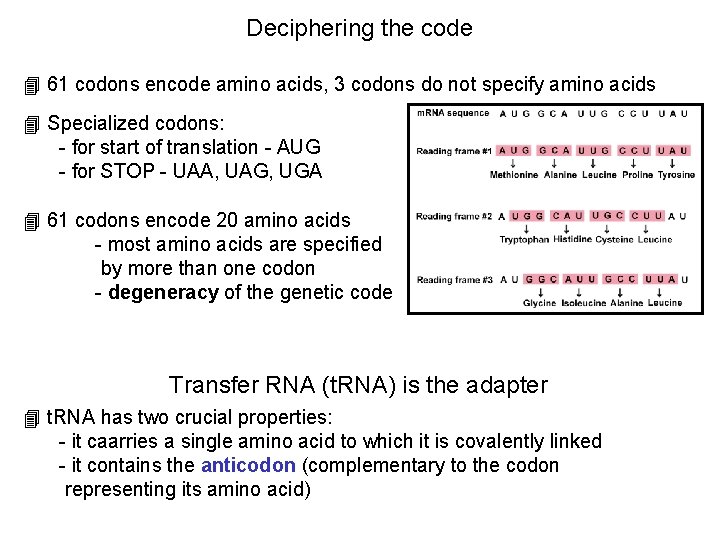
Deciphering the code 4 61 codons encode amino acids, 3 codons do not specify amino acids 4 Specialized codons: - for start of translation - AUG - for STOP - UAA, UAG, UGA 4 61 codons encode 20 amino acids - most amino acids are specified by more than one codon - degeneracy of the genetic code Transfer RNA (t. RNA) is the adapter 4 t. RNA has two crucial properties: - it caarries a single amino acid to which it is covalently linked - it contains the anticodon (complementary to the codon representing its amino acid)

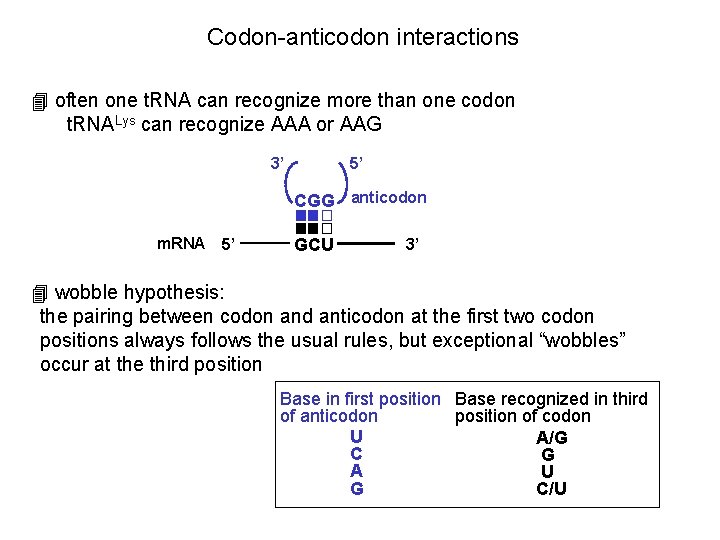
Codon-anticodon interactions 4 often one t. RNA can recognize more than one codon t. RNALys can recognize AAA or AAG 3’ 5’ CGG anticodon m. RNA 5’ GCU 3’ 4 wobble hypothesis: the pairing between codon and anticodon at the first two codon positions always follows the usual rules, but exceptional “wobbles” occur at the third position Base in first position Base recognized in third of anticodon position of codon U A/G C G A U G C/U

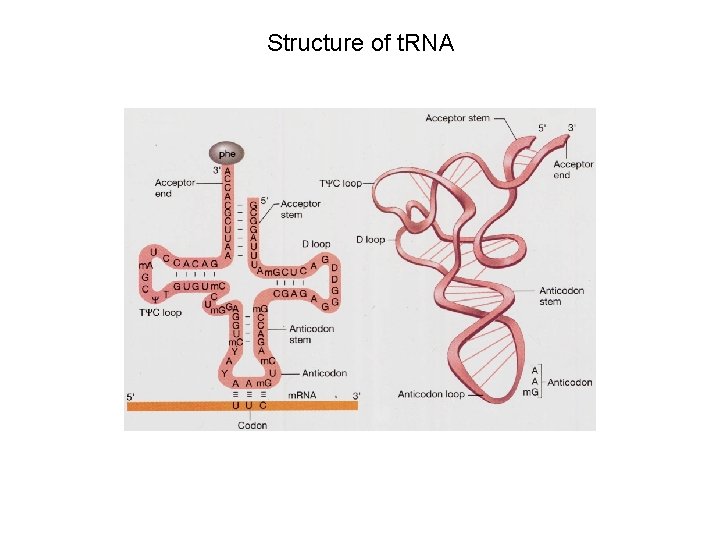
Structure of t. RNA

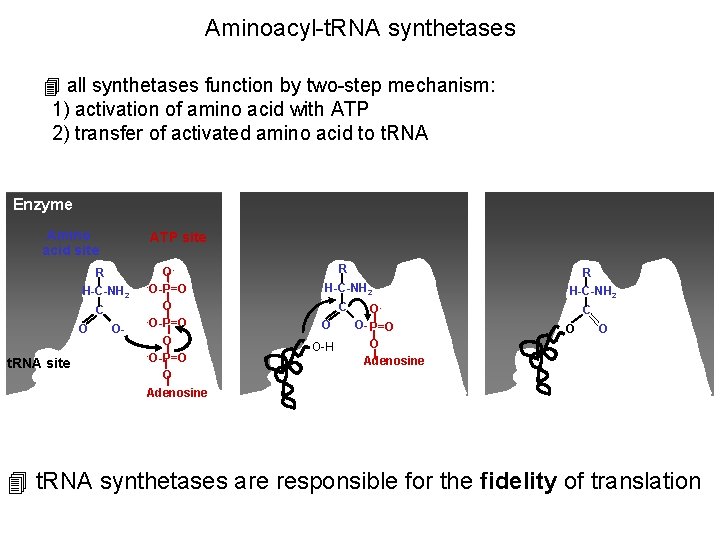
Aminoacyl-t. RNA synthetases 4 all synthetases function by two-step mechanism: 1) activation of amino acid with ATP 2) transfer of activated amino acid to t. RNA Enzyme Amino acid site ATP site R H-C-NH 2 C O t. RNA site O- O-O-P=O O Adenosine R R H-C-NH 2 C O O-H OO- P=O O Adenosine H-C-NH 2 C O O 4 t. RNA synthetases are responsible for the fidelity of translation

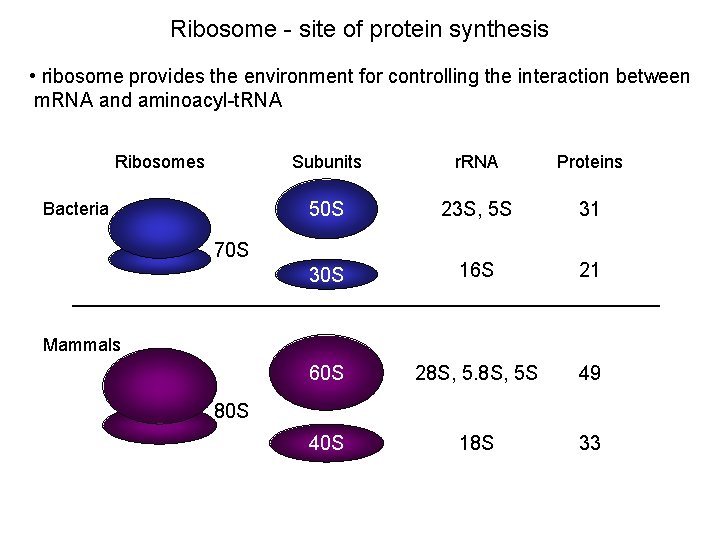
Ribosome - site of protein synthesis • ribosome provides the environment for controlling the interaction between m. RNA and aminoacyl-t. RNA Ribosomes Bacteria Subunits r. RNA Proteins 50 S 23 S, 5 S 31 30 S 16 S 21 60 S 28 S, 5 S 49 40 S 18 S 33 70 S Mammals 80 S

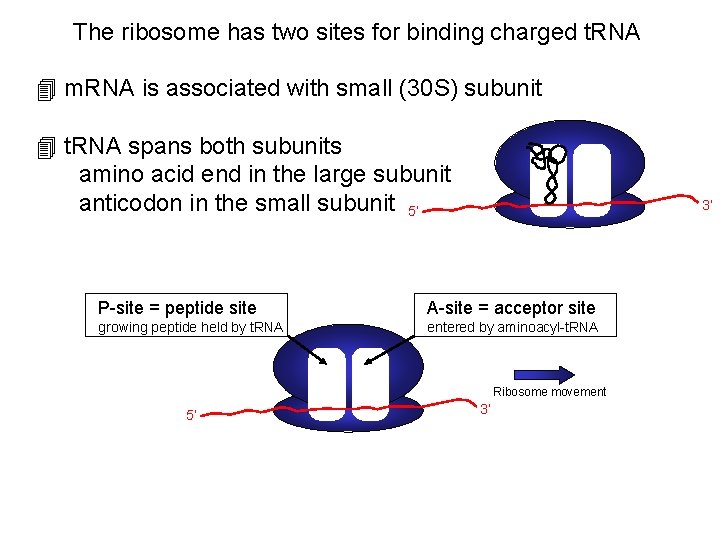
The ribosome has two sites for binding charged t. RNA 4 m. RNA is associated with small (30 S) subunit 4 t. RNA spans both subunits amino acid end in the large subunit anticodon in the small subunit 5’ 3’ P-site = peptide site A-site = acceptor site growing peptide held by t. RNA entered by aminoacyl-t. RNA Ribosome movement 5’ 3’

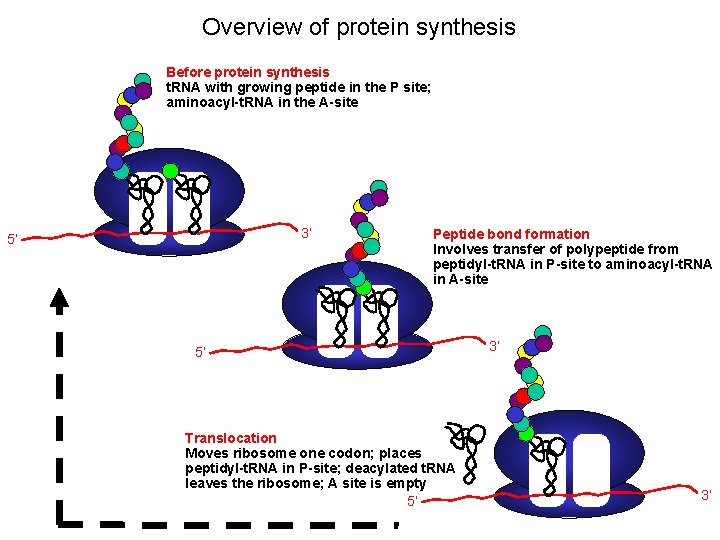
Overview of protein synthesis Before protein synthesis t. RNA with growing peptide in the P site; aminoacyl-t. RNA in the A-site 3’ 5’ Peptide bond formation Involves transfer of polypeptide from peptidyl-t. RNA in P-site to aminoacyl-t. RNA in A-site 5’ Translocation Moves ribosome one codon; places peptidyl-t. RNA in P-site; deacylated t. RNA leaves the ribosome; A site is empty 5’ 3’ 3’

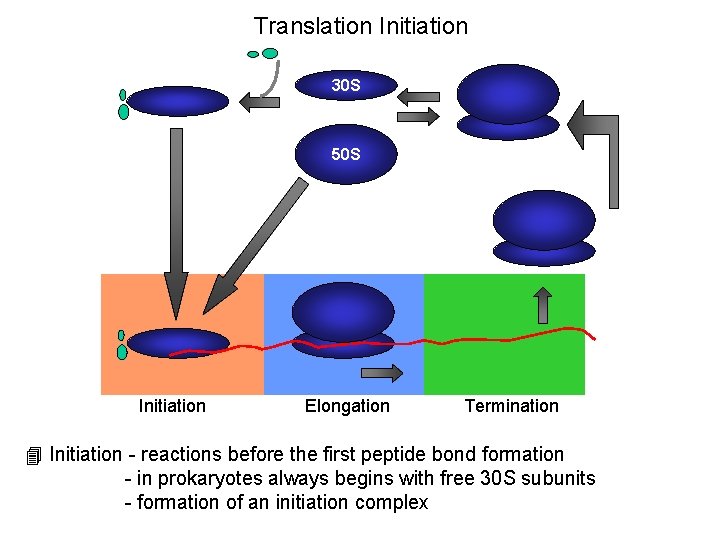
Translation Initiation 30 S 50 S Initiation Elongation Termination 4 Initiation - reactions before the first peptide bond formation - in prokaryotes always begins with free 30 S subunits - formation of an initiation complex

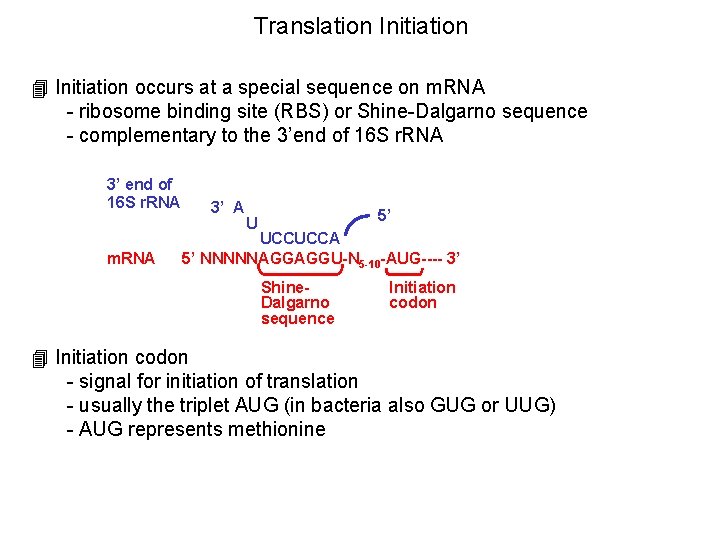
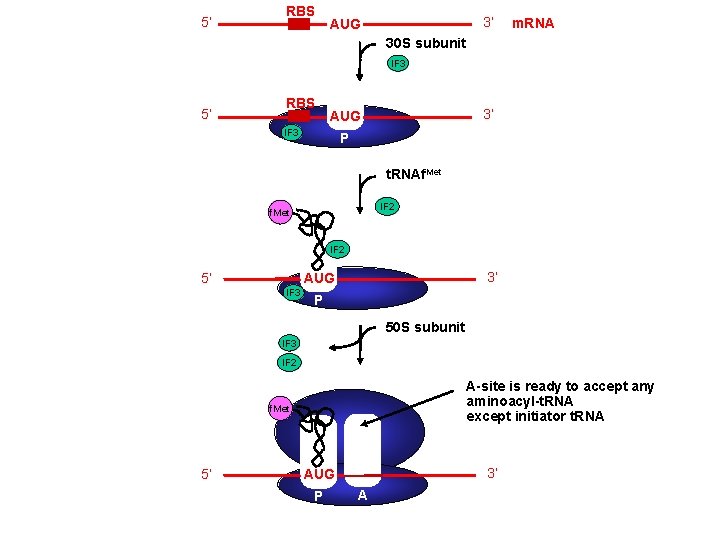
Translation Initiation 4 Initiation occurs at a special sequence on m. RNA - ribosome binding site (RBS) or Shine-Dalgarno sequence - complementary to the 3’end of 16 S r. RNA 3’ end of 16 S r. RNA m. RNA 3’ A 5’ U UCCUCCA 5’ NNNNNAGGAGGU-N 5 -10 -AUG---- 3’ Shine. Dalgarno sequence Initiation codon 4 Initiation codon - signal for initiation of translation - usually the triplet AUG (in bacteria also GUG or UUG) - AUG represents methionine

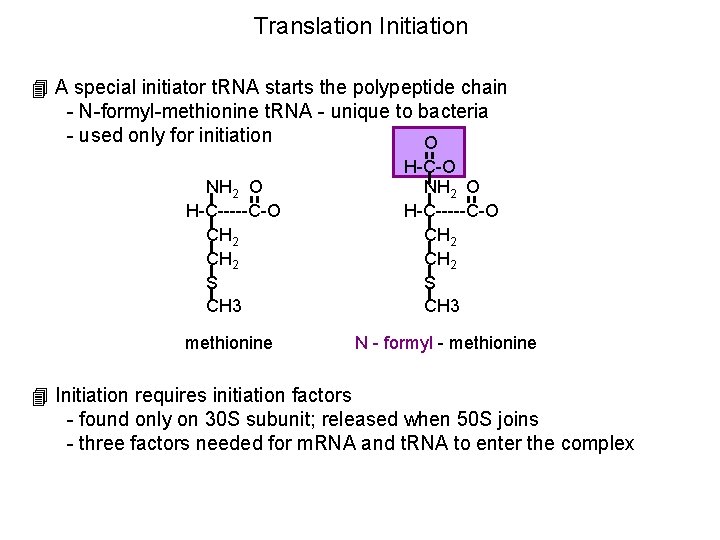
Translation Initiation 4 A special initiator t. RNA starts the polypeptide chain - N-formyl-methionine t. RNA - unique to bacteria - used only for initiation O NH 2 O H-C-----C-O CH 2 S CH 3 H-C-O NH 2 O H-C-----C-O CH 2 S CH 3 methionine N - formyl - methionine 4 Initiation requires initiation factors - found only on 30 S subunit; released when 50 S joins - three factors needed for m. RNA and t. RNA to enter the complex

5’ RBS 3’ AUG m. RNA 30 S subunit IF 3 5’ RBS 3’ AUG IF 3 P t. RNAf. Met IF 2 5’ 3’ AUG IF 3 P 50 S subunit IF 3 IF 2 A-site is ready to accept any aminoacyl-t. RNA except initiator t. RNA f. Met 5’ 3’ AUG P A

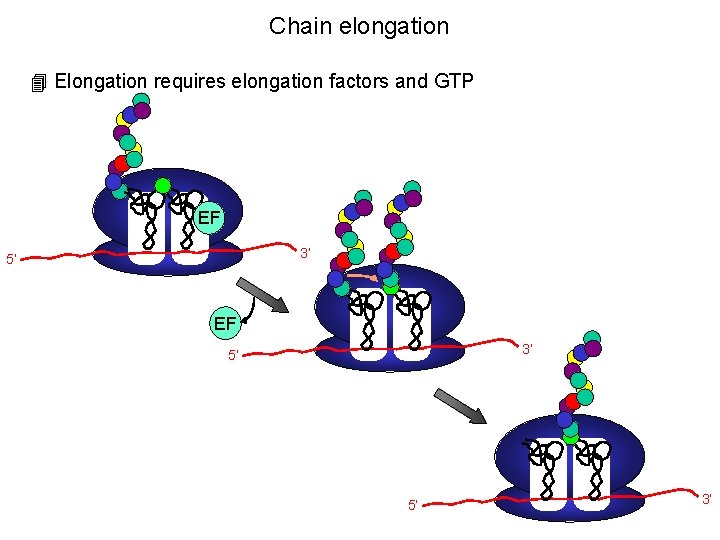
Chain elongation 4 Elongation requires elongation factors and GTP EF 3’ 5’ 5’ 3’

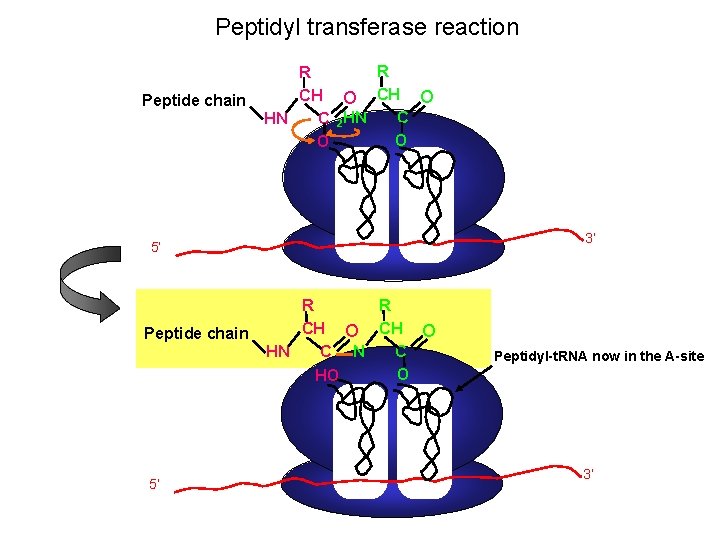
Peptidyl transferase reaction Peptide chain R R CH O C HN C 2 HN O O 3’ 5’ Peptide chain 5’ R R CH O C HN C N O HO Peptidyl-t. RNA now in the A-site 3’

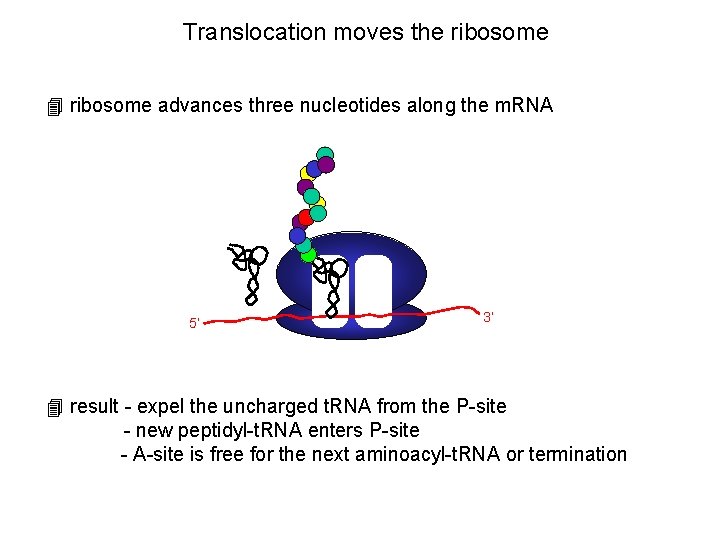
Translocation moves the ribosome 4 ribosome advances three nucleotides along the m. RNA 5’ 3’ 4 result - expel the uncharged t. RNA from the P-site - new peptidyl-t. RNA enters P-site - A-site is free for the next aminoacyl-t. RNA or termination

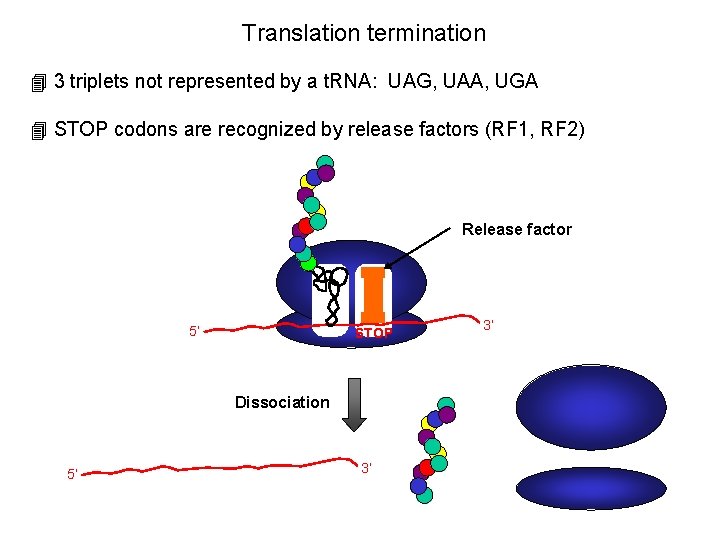
Translation termination 4 3 triplets not represented by a t. RNA: UAG, UAA, UGA 4 STOP codons are recognized by release factors (RF 1, RF 2) Release factor 5’ STOP Dissociation 5’ 3’ 3’

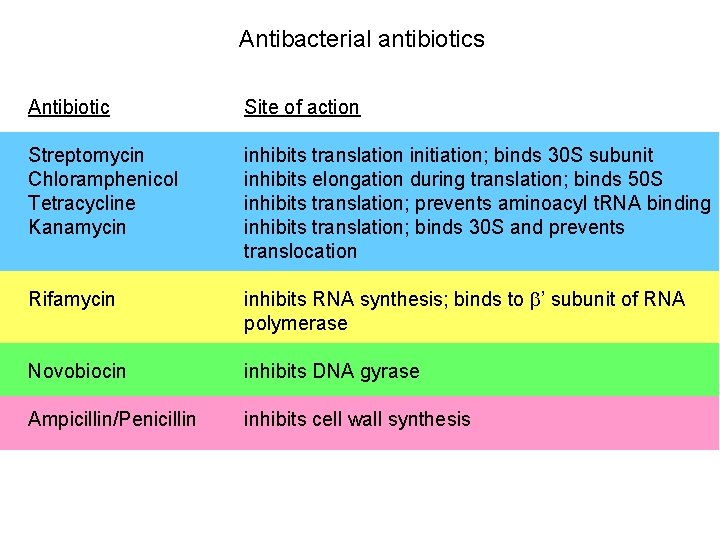
Antibacterial antibiotics Antibiotic Site of action Streptomycin Chloramphenicol Tetracycline Kanamycin inhibits translation initiation; binds 30 S subunit inhibits elongation during translation; binds 50 S inhibits translation; prevents aminoacyl t. RNA binding inhibits translation; binds 30 S and prevents translocation Rifamycin inhibits RNA synthesis; binds to b’ subunit of RNA polymerase Novobiocin inhibits DNA gyrase Ampicillin/Penicillin inhibits cell wall synthesis

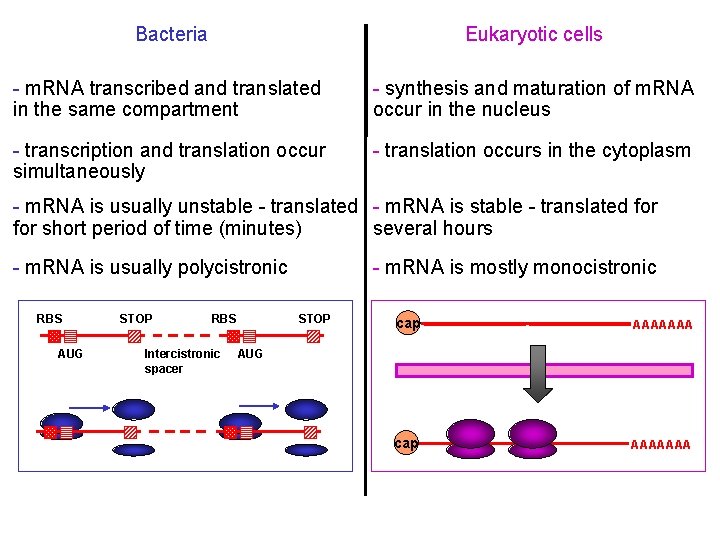
Bacteria Eukaryotic cells - m. RNA transcribed and translated in the same compartment - synthesis and maturation of m. RNA occur in the nucleus - transcription and translation occur simultaneously - translation occurs in the cytoplasm - m. RNA is usually unstable - translated - m. RNA is stable - translated for short period of time (minutes) several hours - m. RNA is usually polycistronic RBS AUG STOP RBS Intercistronic spacer - m. RNA is mostly monocistronic STOP cap AAAAAAA AUG

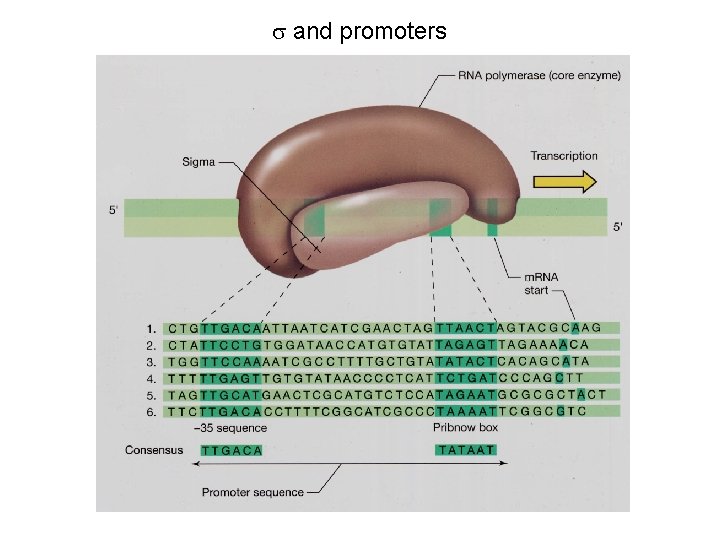
s and promoters

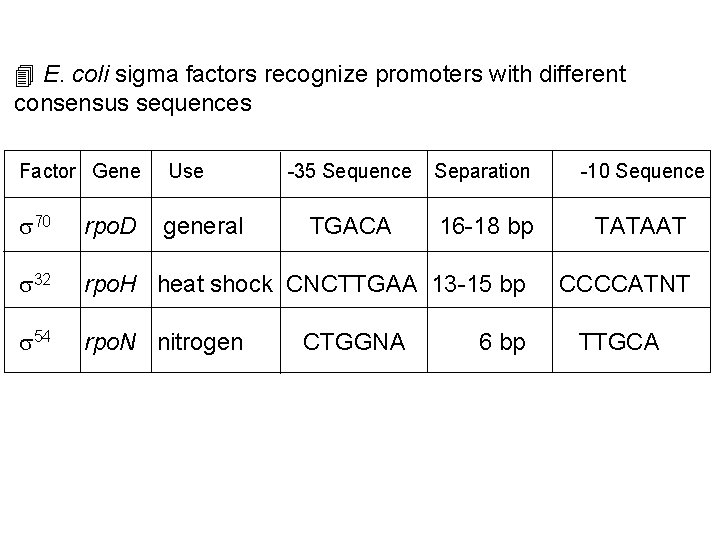
4 E. coli sigma factors recognize promoters with different consensus sequences Factor Gene Use -35 Sequence Separation -10 Sequence s 70 rpo. D general TGACA 16 -18 bp TATAAT s 32 rpo. H heat shock CNCTTGAA 13 -15 bp s 54 rpo. N nitrogen CTGGNA 6 bp CCCCATNT TTGCA