THE GENETIC CODE Genes contain the coded formula














- Slides: 26

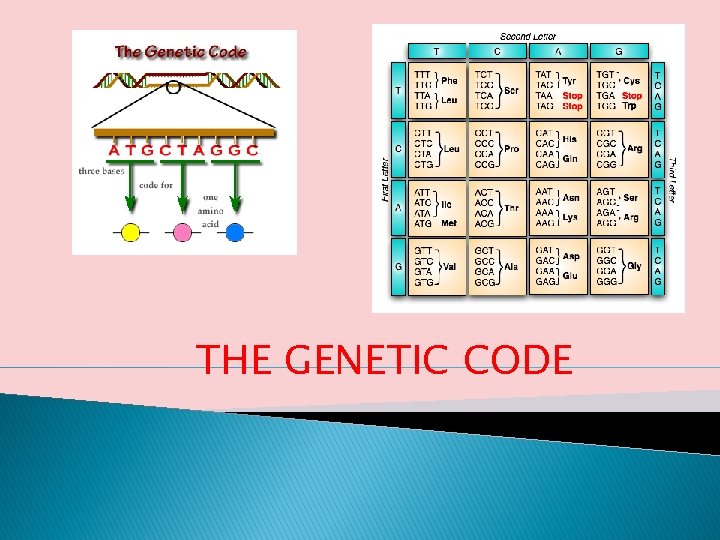
THE GENETIC CODE

� Genes contain the coded formula needed by the cell to produce proteins. � The sequence of nucleotides in a typical gene is a genetic code that carries the information for making an RNA � The term genetic code refers to the set of three-base code words (codons) in m. RNAs that stand for the 20 amino acids in proteins. A DNA molecule is composed of three kinds of moieties: (i) phosphoric acid, (ii) deoxyribose sugar, and (iii) nitrogen bases. �

1. The code is a triplet codon � The colinearity of protein molecules and DNA polynucleotides has given the clue that the specific arrangement of four nitrogen bases (e. g. , A, T, C and G) in DNA polynucleotide chains, somehow, determines the sequence of amino acids in protein molecules. Therefore, these four DNA bases can be considered as four alphabets of DNA molecule. All the genetic informations, therefore, should be written by these four alphabets of DNA. Now the question

� By the 1960 s it had long been apparent that at least three nucleotide residues of DNA are necessary to encode each amino acid. � The four code letters of DNA (A, T, G, and C) in groups of two can yield only 42= 16 different combinations, insufficient to encode 20 aminoacids. � Groups of three, however, yield 43= 64 different combinations.

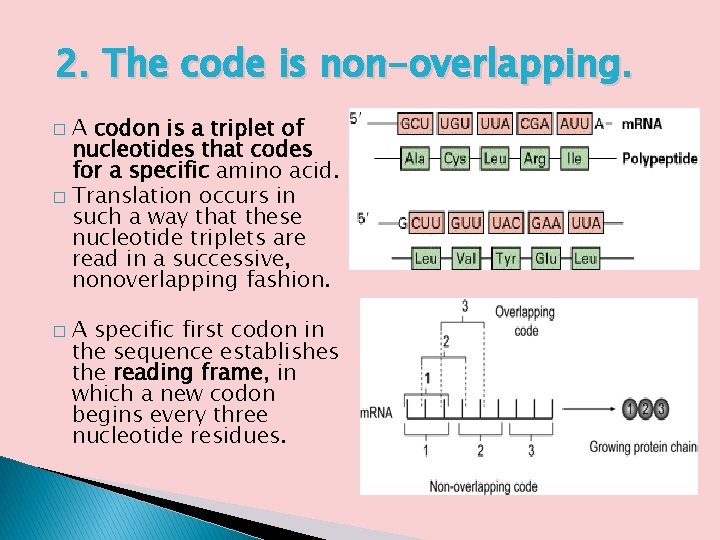
2. The code is non-overlapping. A codon is a triplet of nucleotides that codes for a specific amino acid. � Translation occurs in such a way that these nucleotide triplets are read in a successive, nonoverlapping fashion. � � A specific first codon in the sequence establishes the reading frame, in which a new codon begins every three nucleotide residues.

Nonoverlapping Codons � � � In a nonoverlapping code, each base is part of at most one codon. In an overlapping code, one base may be part of two or even three codons. Consider the following micromessage: AUGUUC Assuming that the code is triplet (three bases per codon) and this message is read from the beginning, the codons will be AUG and UUC if the code is nonoverlapping. On the other hand, an overlapping code might yield four codons: AUG, UGU, GUU, and UUC. As early as 1957, Sydney Brenner concluded on theoretical grounds that a fully overlapping triplet code like this would be impossible.

Nonoverlapping Codons However, given the data available in 1957, a partially overlapping code remained possible, but A. Tsugita and H. Frankel-Conrat laid it to rest with the following line of reasoning: � If the code is nonoverlapping, a change of one base in an m. RNA (a missense mutation) would change no more than one amino acid in the resulting protein. � For example, consider another micromessage: � AUGCUA � Assuming that the code is triplet (three bases per codon) and this message is read from the beginning, the codons will be AUG and CUA if the code is nonoverlapping

Nonoverlapping Codons �A change in the third base (G) would change only one codon (AUG) and therefore at most only one amino acid. � On the other hand, if the code were overlapping, base G could be part of three adjacent codons (AUG, UGC, and GCU). � Therefore, if the G were changed, up to three adjacent amino acids could be changed in the resulting protein. � But when the investigators introduced one-base alterations into m. RNA from tobacco mosaic virus (TMV), they found that these never caused changes in more than one amino acid. � Hence, the code must be nonoverlapping.

3. The code is commaless. � If the code contained untranslated gaps, or “commas, ” mutations that add or subtract a base from a message might change a only few codons. � In other words, these mutations might frequently be lethal, but in many cases the mutation should occur just before a comma in the message and therefore have little, if any, effect. � If no commas were present to get the ribosome back on track, these mutations would be lethal except when they occur right at the end of a message.

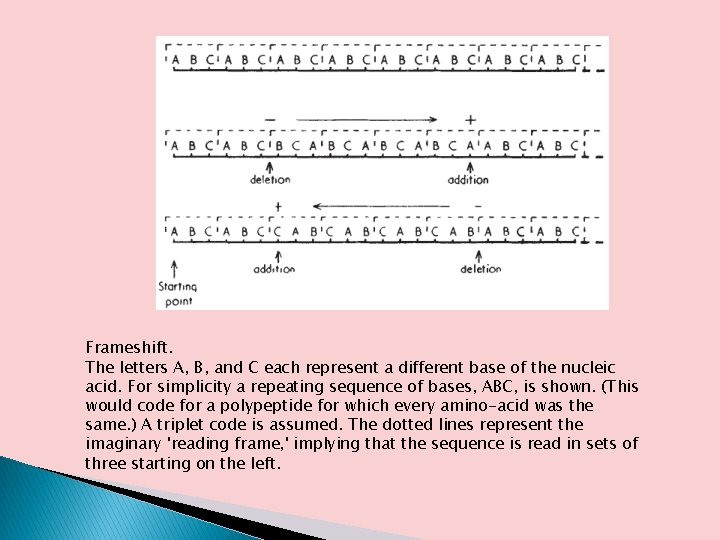
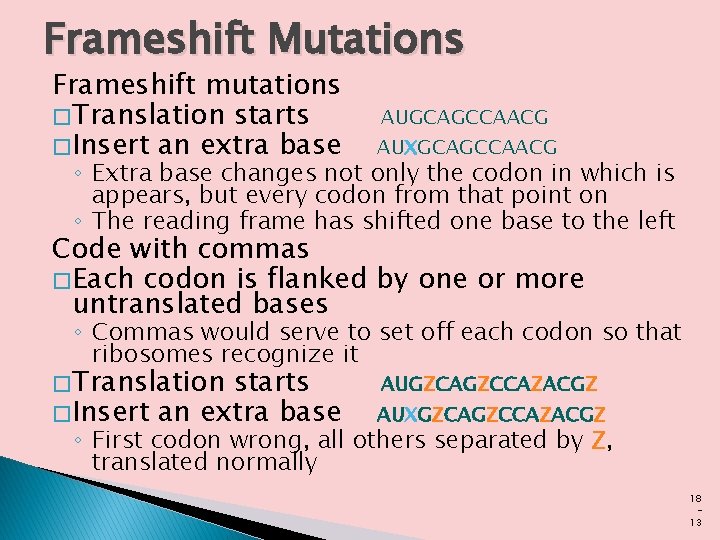
No Gaps in the Code � � � Such mutations do occur, and they are called frame shift mutations; they work as follows. Consider another tiny message: AUGCAGCCAACG If translation starts at the beginning, the codons will be AUG, CAG, CCA, and ACG. If we insert an extra base (X) right after base U, we get: AUXGCAGCCAACG Now this would be translated from the beginning as AUX, GCA, GCC, AAC. Notice that the extra base changes not only the codon (AUX) in which it appears, but every codon from that point on. The reading frame has shifted one base to the left; whereas C was originally the first base of the second codon, G is now in that position. �

No Gaps in the Code On the other hand, a code with commas would be one in which each codon is flanked by one or more untranslated bases, represented by Z’s in the following message. � The commas would serve to set off each codon so the ribosome could recognize it: AUGZCAGZCCAZACGZ � Deletion or insertion of a base anywhere in this message would change only a single codon. � The comma (Z) at the end of the damaged codon would then put the ribosome back on the right track. � Thus, addition of an extra base (X) to the first codon would give the message: AUXGZCAGZCCAZACGZ � The first codon (AUXG) is now wrong, but all the others, still neatly set off by Z’s, would be translated normally �

Frameshift. The letters A, B, and C each represent a different base of the nucleic acid. For simplicity a repeating sequence of bases, ABC, is shown. (This would code for a polypeptide for which every amino-acid was the same. ) A triplet code is assumed. The dotted lines represent the imaginary 'reading frame, ' implying that the sequence is read in sets of three starting on the left.

Frameshift Mutations Frameshift mutations � Translation starts � Insert an extra base AUGCAGCCAACG AUXGCAGCCAACG ◦ Extra base changes not only the codon in which is appears, but every codon from that point on ◦ The reading frame has shifted one base to the left Code with commas � Each codon is flanked by one or more untranslated bases ◦ Commas would serve to set off each codon so that ribosomes recognize it � Translation starts � Insert an extra base AUGZCAGZCCAZACGZ AUXGZCAGZCCAZACGZ ◦ First codon wrong, all others separated by Z, translated normally 18 13

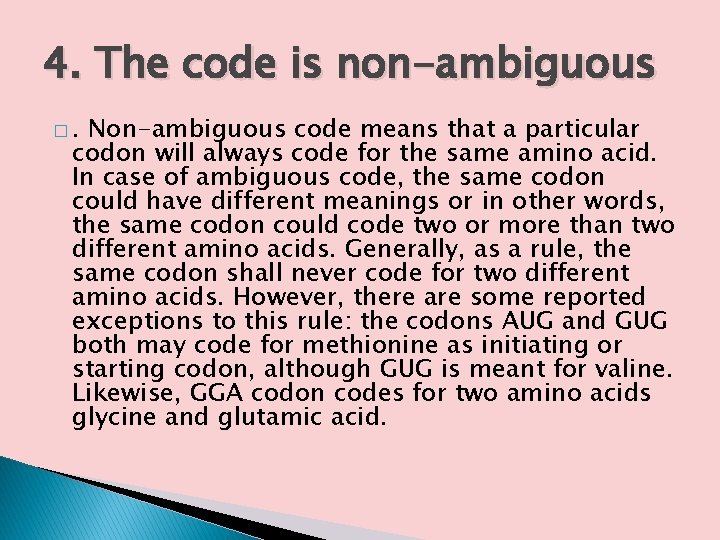
4. The code is non-ambiguous �. Non-ambiguous code means that a particular codon will always code for the same amino acid. In case of ambiguous code, the same codon could have different meanings or in other words, the same codon could code two or more than two different amino acids. Generally, as a rule, the same codon shall never code for two different amino acids. However, there are some reported exceptions to this rule: the codons AUG and GUG both may code for methionine as initiating or starting codon, although GUG is meant for valine. Likewise, GGA codon codes for two amino acids glycine and glutamic acid.

5. The code has polarity. � THe code is always read in a fixed direction, i. e. , in the 5'→ 3' direction. In other words, the codon has a polarity.

6. The code is degenerate �. More than one codon may specify the same amino acid; this is called degeneracy of the code. For example, except for tryptophan and methionine, which have a single codon each, all other 18 amino acids have more than one codon. Thus, nine amino acids, namely phenylalanine, tyrosine, histidine, glutamine, asparagine, lysine, aspartic acid, glutamic acid and cysteine, have two codons each. Isoleucine has three codons. Five amino acids, namely valine, proline, threonine, alanine and glycine, have four codons each. Three amino acids, namely leucine, arginine and serine, have six codons eac

� The initiation codon AUG is the most common signal for the beginning of a polypeptide in all cells � In addition to coding for Met residues in internal positions of polypeptides. � The termination codons (UAA, UAG, and UGA), also called stop codons or nonsense codons


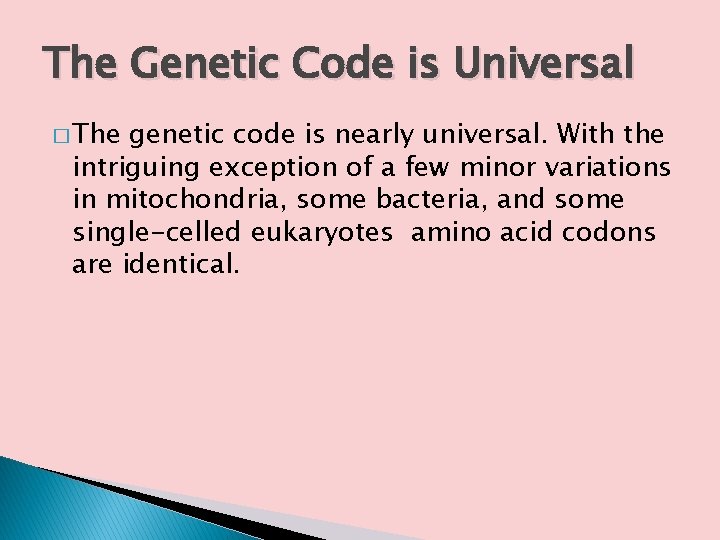
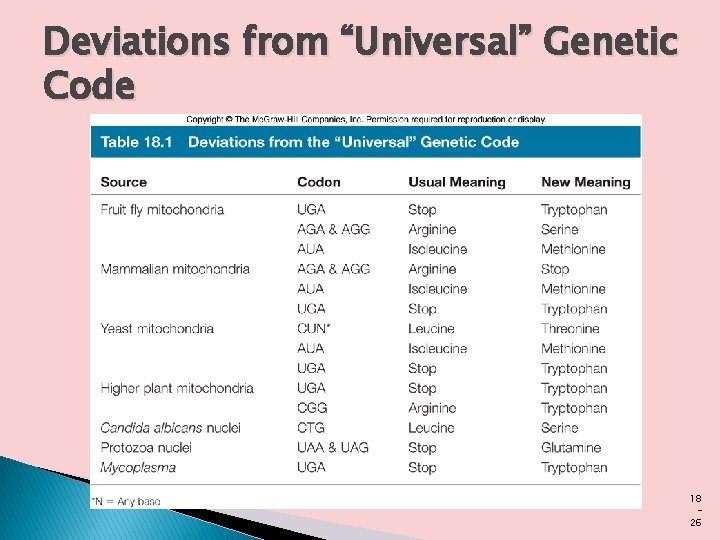
The Genetic Code is Universal � The genetic code is nearly universal. With the intriguing exception of a few minor variations in mitochondria, some bacteria, and some single-celled eukaryotes amino acid codons are identical.

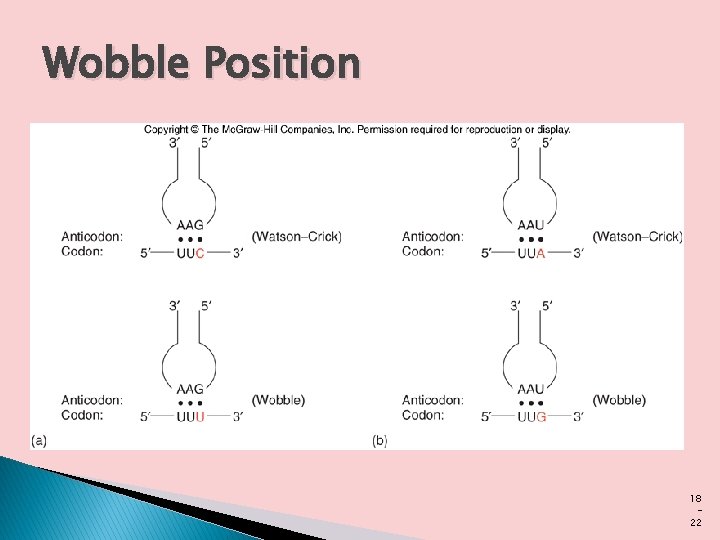
Wobble Hypothesis � When several different codons specify one amino acid, the difference between them usually lies at the third base position (at the 3 end). � For example, alanine is coded by the triplets GCU, GCC, GCA, and GCG. � The codons for most amino acids can be symbolized by XYAG or XYUC �. The first two letters of each codon are the primary determinants of specificity, a feature that has some interesting consequences.

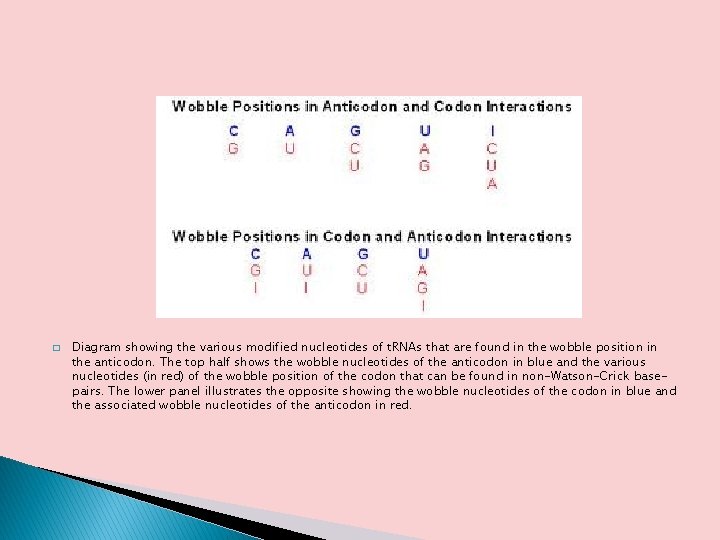
� Diagram showing the various modified nucleotides of t. RNAs that are found in the wobble position in the anticodon. The top half shows the wobble nucleotides of the anticodon in blue and the various nucleotides (in red) of the wobble position of the codon that can be found in non-Watson-Crick basepairs. The lower panel illustrates the opposite showing the wobble nucleotides of the codon in blue and the associated wobble nucleotides of the anticodon in red.

Wobble Position 18 22


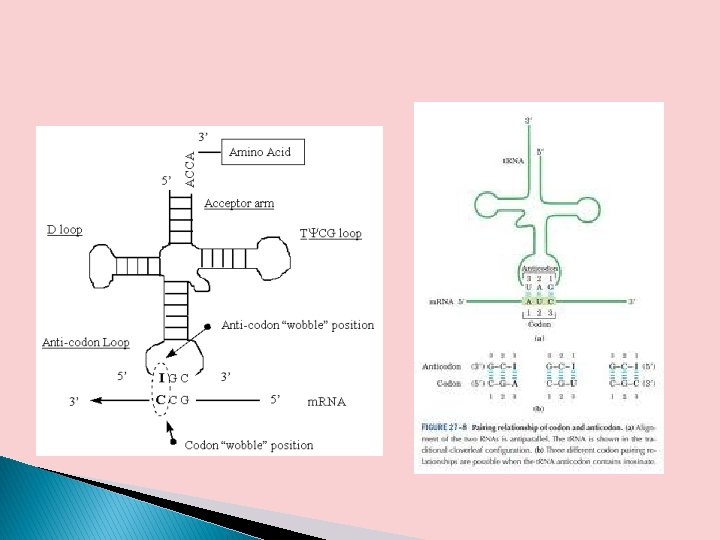
� Crick proposed a set of four relationships called the wobble hypothesis 1. The first two bases of an m. RNA codon always form strong Watson-Crick base pairs with the corresponding bases of the t. RNA anticodon and confer most of the coding specificity. � 2. The first base of the anticodon (reading in the 5’→ 3’ � � � direction; this pairs with the third base of the codon) determines the number of codons recognized by the t. RNA. When the first base of the anticodon is C or A, base pairing is specific and only one codon is recognized by that t. RNA. When the first base is U or G, binding is less specific and two different codons may be read. When inosine (I) is the first (wobble) nucleotide of an anticodon, three different codons can be recognized—the maximum number for any t. RNA.

� 3. When an amino acid is specified by several different codons, the codons that differ in eitherof the first two bases require different t. RNAs. � 4. A minimum of 32 t. RNAs are required to translate all 61 codons (31 to encode the amino acids and 1 for initiation).

Deviations from “Universal” Genetic Code 18 26