The Generation Challenge Programme GCP Platform for Crop








- Slides: 35

The Generation Challenge Programme (GCP) Platform for Crop Research Richard Bruskiewich and the rest of … 9/29/2020 1

…The GCP SP 4 team and Contributors Theo van Hintum (WUR), GCP Subprogramme 4 Leader IRRI-CIMMYT Crop Research Informatics Laboratory CIP: University of British Columbia: Alexis Dereeper Reinhard Simon Mark Wilkinson Matthieu Conte Edwin Rojas Brigitte Courtois ICRISAT: GSC Bioinformatics Graduate Program, BC Cancer Agency: CIRAD: Manuel Ruiz Graham Mc. Laren Guy Davenport Bioversity: Jayashree Balaji Thomas Metz Trushar Shah Mathieu Rouard ICARDA: Martin Senger Kyle Braak Tom Hazekamp Akinnola Akintunde Ramil Mauleon Sebastian Ritter Milko Skofic NCGR: Mylah Anacleto Raj Sood Andrew Farmer Michael Jonathan Mendoza NIAS: Gary Schiltz Victor Jun Ulat Yi Zhang Masaru Takeya SCRI: Arllet Portugal Sergio Gregorio Koji Doi Jennifer Lee Ryan Alamban Joseph Hermocilla Kouji Satoh David Marshall Lord Hendrix Barboza Michael Echavez Jeffrey Detras Roque Almodiel Shoshi Kikuchi Cornell University: EMBRAPA: Terry Casstevens Kevin Manansala Marcos Costa Pankaj Jaiswal Jeffrey Morales Natalia Martins Dave Matthews Georgios Pappas ACGT: Barry Peralta Samart Wanchana Rowena Valerio Supat Thongjuea Nelzo Ereful Ayton Meintjes Jane Morris Benjamin Good James Wagner

Overview n Generation Challenge Programme crop informatics research and development n GCP platform architecture: q Domain model & ontology q Application development framework

Challenge Programme “I challenge the next generation to use new scientific tools and techniques to address the problems that plague the world’s poor” Dr. Norman Borlaug http: //www. generationcp. org

What is it? n n n An international research programme established in 2003, projected to last 10 years, and hosted by the CGIAR with global partners from ARI and NARES Research Themes Directed to Crop Improvement: q Genomics and comparative biology across species q Characterization of genetic diversity for allele mining q Gene transfer technologies Five research subprogrammes, one of which is crop information systems development.

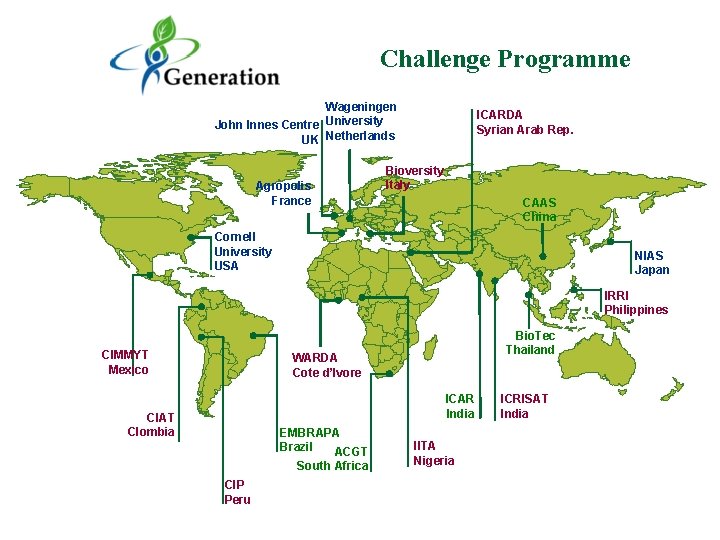
Challenge Programme Wageningen John Innes Centre University UK Netherlands Agropolis France ICARDA Syrian Arab Rep. Bioversity Italy CAAS China Cornell University USA NIAS Japan IRRI Philippines CIMMYT Mexico Bio. Tec Thailand WARDA Cote d’Ivore ICAR India CIAT Clombia EMBRAPA Brazil ACGT South Africa CIP Peru IITA Nigeria ICRISAT India

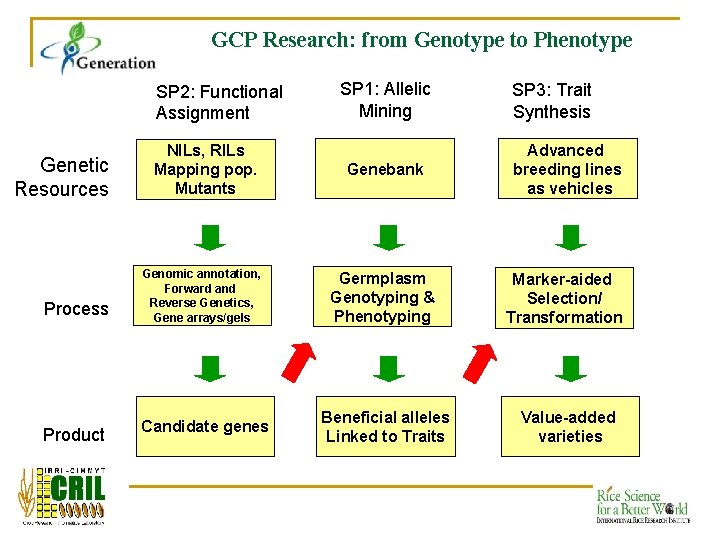
GCP Research: from Genotype to Phenotype SP 2: Functional Assignment Genetic Resources Process Product SP 1: Allelic Mining SP 3: Trait Synthesis NILs, RILs Mapping pop. Mutants Genebank Advanced breeding lines as vehicles Genomic annotation, Forward and Reverse Genetics, Gene arrays/gels Germplasm Genotyping & Phenotyping Marker-aided Selection/ Transformation Candidate genes Beneficial alleles Linked to Traits Value-added varieties

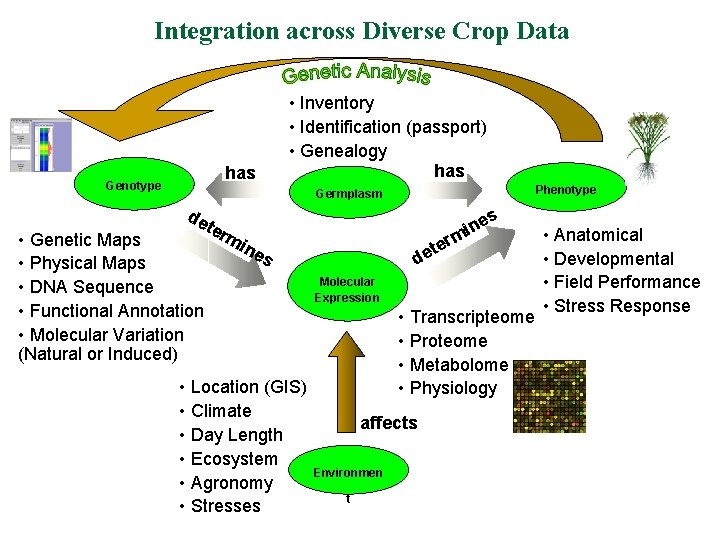
Integration across Diverse Crop Data • Inventory • Identification (passport) • Genealogy has Genotype Phenotype Germplasm de ter • Genetic Maps • Physical Maps • DNA Sequence • Functional Annotation • Molecular Variation (Natural or Induced) mi s ne i m r e t de ne s • Location (GIS) • Climate • Day Length • Ecosystem • Agronomy • Stresses Molecular Expression • Transcripteome • Proteome • Metabolome • Physiology affects Environmen t • Anatomical • Developmental • Field Performance • Stress Response

Crop Information Systems: the Next n Large, globally distributed consortium n Diverse research requiring a diversity of tools n Large data sets with diverse data types n Many legacy informatics systems and tools n Global data integration required… Key Issue: Interoperability

Some Basic GCP Research Objectives n Compile a list of germplasm meeting specific passport data criteria n Compile a list of genetic markers of interest from genetic and QTL maps n Retrieve genotypes of specified markers, for specified germplasm n Align gene expression data against QTL positional evidence to identify candidate gene loci for specified traits

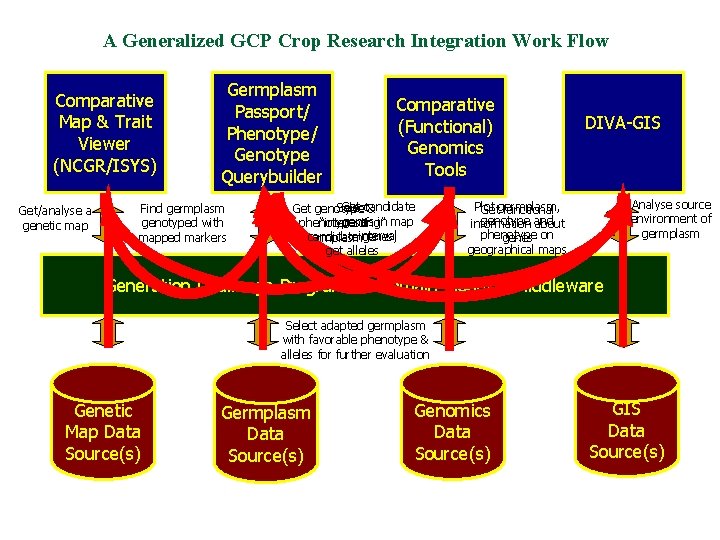
A Generalized GCP Crop Research Integration Work Flow Comparative Map & Trait Viewer (NCGR/ISYS) Get/analyse a genetic map Germplasm Passport/ Phenotype/ Genotype Querybuilder Find germplasm genotyped with mapped markers Comparative (Functional) Genomics Tools Get candidate Select Get genotype & genes “interesting” phenotype of in map candidateinterval genes; germplasm get alleles DIVA-GIS Analyse source environment of germplasm Plot Getgermplasm, functional genotype and information about phenotype genes on geographical maps Generation Challenge Programme Domain Model & Middleware Select adapted germplasm with favorable phenotype & alleles for further evaluation Genetic Map Data Source(s) Germplasm Data Source(s) Genomics Data Source(s) GIS Data Source(s)

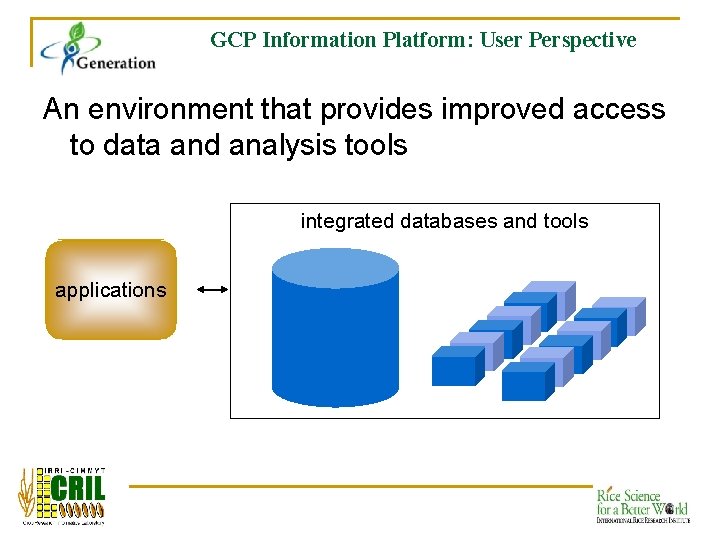
GCP Information Platform: User Perspective An environment that provides improved access to data and analysis tools integrated databases and tools applications

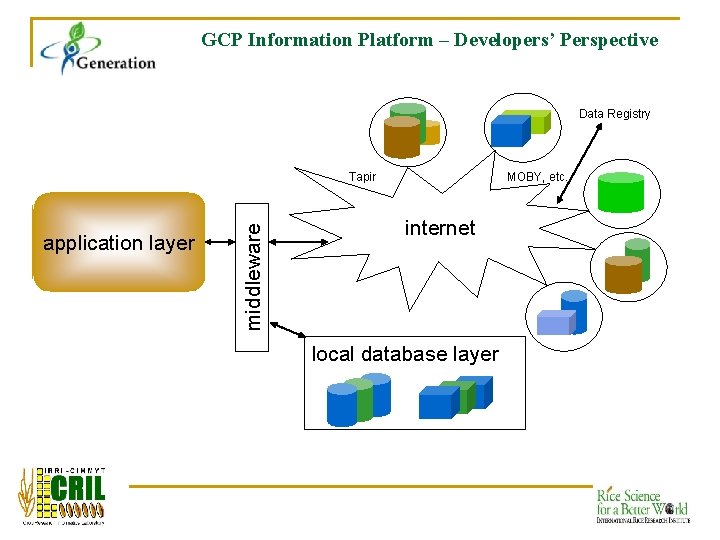
GCP Information Platform – Developers’ Perspective Data Registry application layer middleware Tapir MOBY, etc. internet local database layer

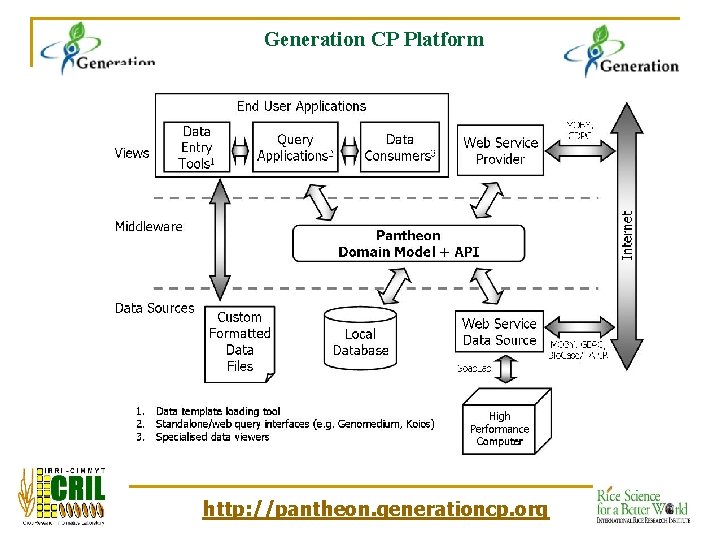
Generation CP Platform http: //pantheon. generationcp. org

GCP Platform - General Architecture n “Model Driven Architecture” based on “platform independent” GCP scientific domain models, parameterized with controlled vocabulary (“ontology”) n GCP domain models mapped onto platform specific implementations. n Reference (Java) GCP platform application programming interface (API)

Semantics of the GCP Model Driven Architecture n GCP is trying to model the meaning (“semantics”) of the crop research world. n Semantics is found in the domain model at three distinct but interconnected levels: q q q System architectural level: general scientific semantics in terms of high-level object concepts (“object types”) and their global inter-relationships. Entity level: attributes and behaviors internal to high-level object types. Attribute level: attribute values of objects that range over data types: simple (e. g. identifiers, numbers), complex (other classes of entities) or ontology (such as Gene Ontology (GO) terms, for a gene product).

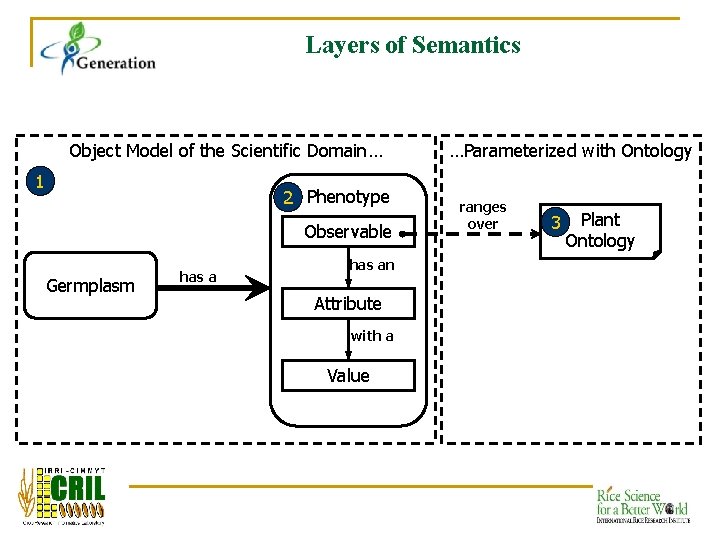
Layers of Semantics Object Model of the Scientific Domain… 1 2 Phenotype Observable Germplasm has an Attribute with a Value …Parameterized with Ontology ranges over 3 Plant Ontology

GCP Domain Model Specification n High-level object types are specified with Unified Modeling Language (UML) and associated text narratives. n Major object classes are represented in the object model. More specialized object types are specified by subclassing major object types using ontology. n Reference model is coded by Eclipse Modeling Language managed with source code versioning and automatically compiled into other representations. http: //pantheon. generationcp. org/demeter

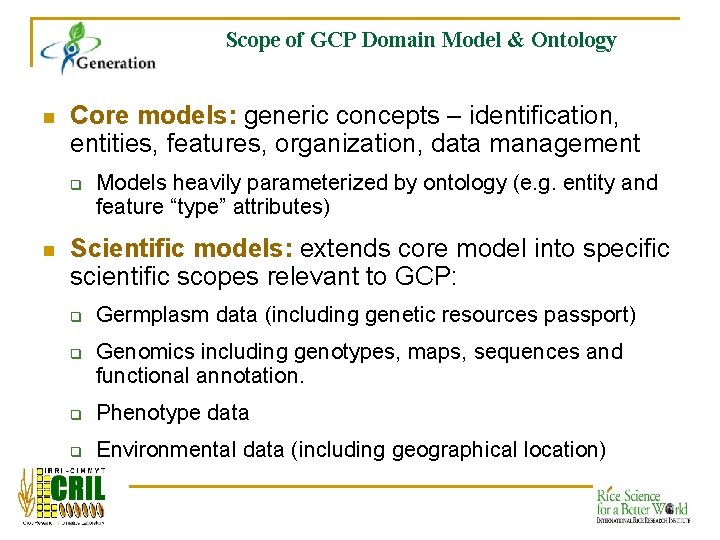
Scope of GCP Domain Model & Ontology n Core models: generic concepts – identification, entities, features, organization, data management q n Models heavily parameterized by ontology (e. g. entity and feature “type” attributes) Scientific models: extends core model into specific scientific scopes relevant to GCP: q q Germplasm data (including genetic resources passport) Genomics including genotypes, maps, sequences and functional annotation. q Phenotype data q Environmental data (including geographical location)

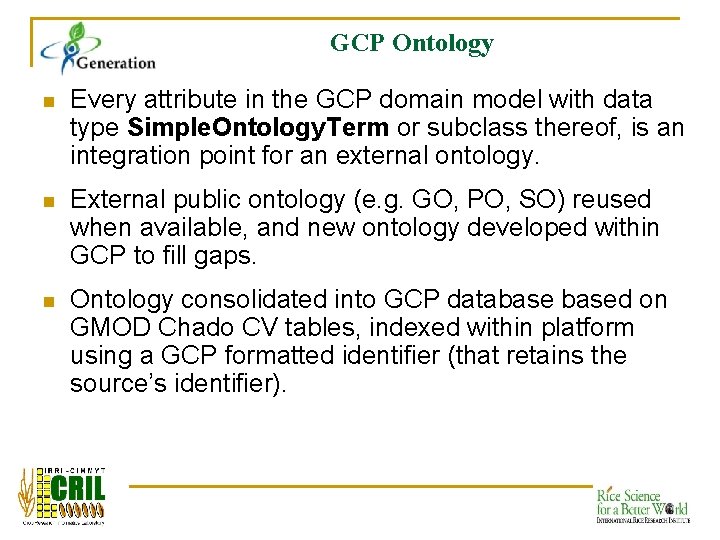
GCP Ontology n Every attribute in the GCP domain model with data type Simple. Ontology. Term or subclass thereof, is an integration point for an external ontology. n External public ontology (e. g. GO, PO, SO) reused when available, and new ontology developed within GCP to fill gaps. n Ontology consolidated into GCP databased on GMOD Chado CV tables, indexed within platform using a GCP formatted identifier (that retains the source’s identifier).

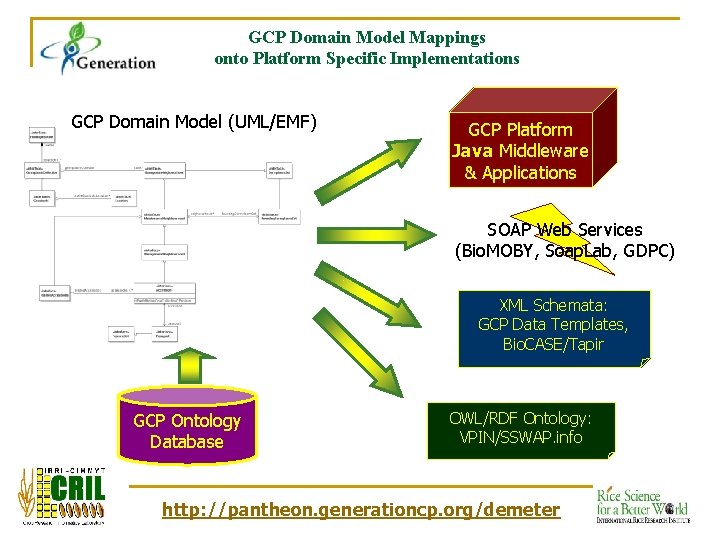
GCP Domain Model Mappings onto Platform Specific Implementations GCP Domain Model (UML/EMF) GCP Platform Java Middleware & Applications SOAP Web Services (Bio. MOBY, Soap. Lab, GDPC) XML Schemata: GCP Data Templates, Bio. CASE/Tapir GCP Ontology Database OWL/RDF Ontology: VPIN/SSWAP. info http: //pantheon. generationcp. org/demeter

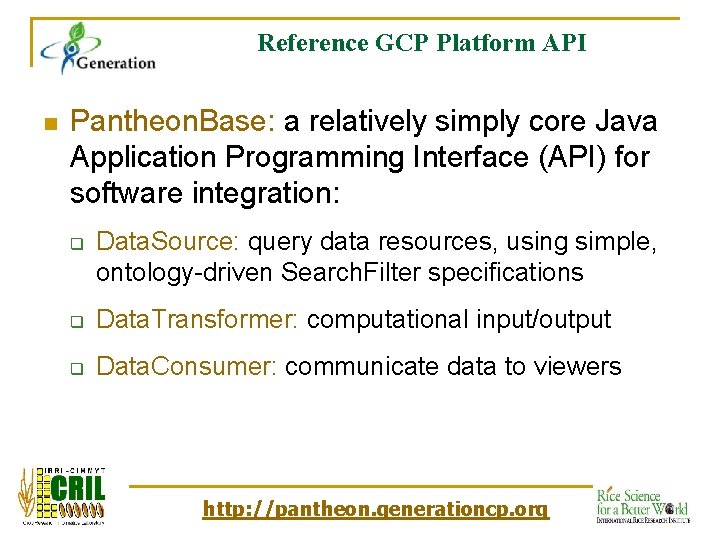
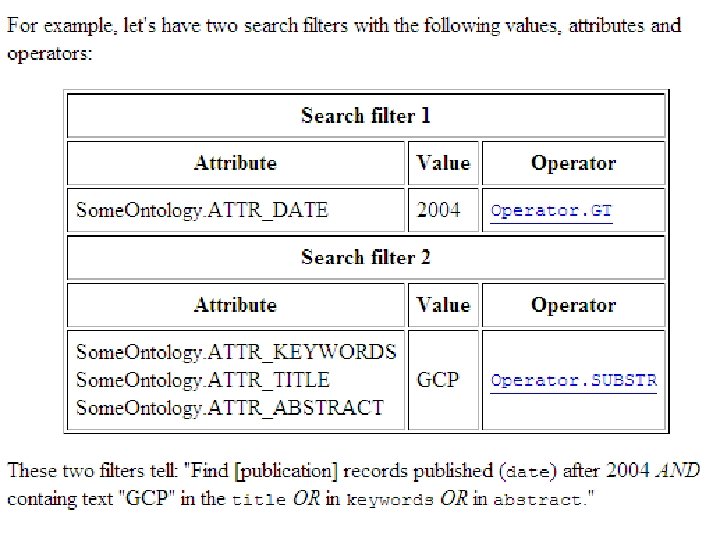
Reference GCP Platform API n Pantheon. Base: a relatively simply core Java Application Programming Interface (API) for software integration: q Data. Source: query data resources, using simple, ontology-driven Search. Filter specifications q Data. Transformer: computational input/output q Data. Consumer: communicate data to viewers http: //pantheon. generationcp. org

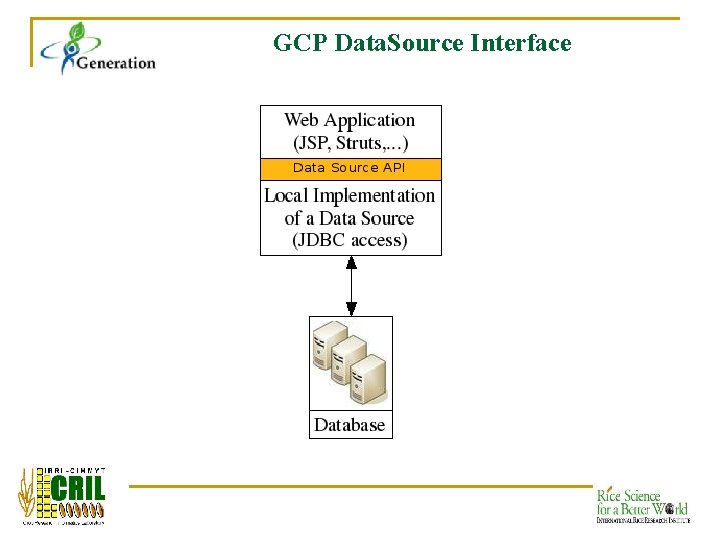
GCP Data. Source Interface

Data. Source Interface

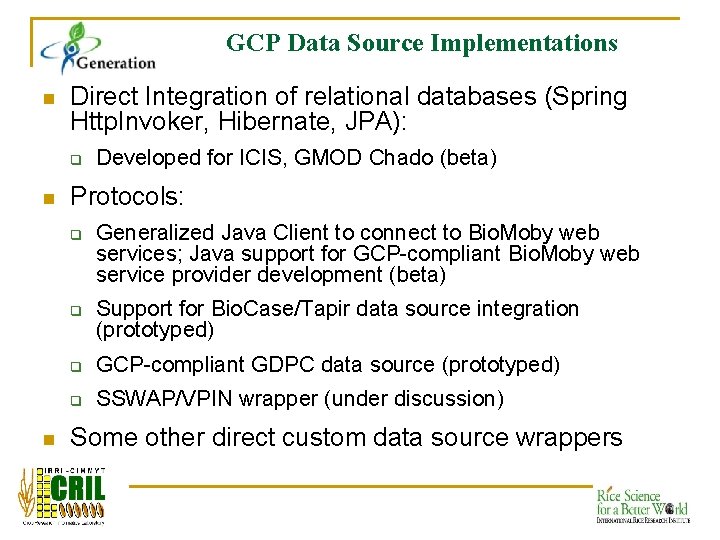
GCP Data Source Implementations n Direct Integration of relational databases (Spring Http. Invoker, Hibernate, JPA): q n Protocols: q q n Developed for ICIS, GMOD Chado (beta) Generalized Java Client to connect to Bio. Moby web services; Java support for GCP-compliant Bio. Moby web service provider development (beta) Support for Bio. Case/Tapir data source integration (prototyped) q GCP-compliant GDPC data source (prototyped) q SSWAP/VPIN wrapper (under discussion) Some other direct custom data source wrappers

Some GCP Bio. MOBY docs… http: //moby. generationcp. org http: //pantheon. generationcp. org/moby http: //cropwiki. irri. org/gcp/index. php/MOBY_Rice_Network

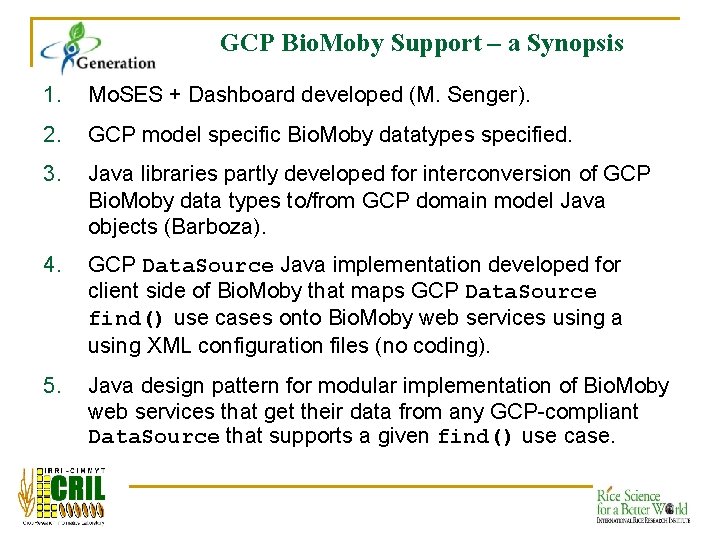
GCP Bio. Moby Support – a Synopsis 1. Mo. SES + Dashboard developed (M. Senger). 2. GCP model specific Bio. Moby datatypes specified. 3. Java libraries partly developed for interconversion of GCP Bio. Moby data types to/from GCP domain model Java objects (Barboza). 4. GCP Data. Source Java implementation developed for client side of Bio. Moby that maps GCP Data. Source find() use cases onto Bio. Moby web services using a using XML configuration files (no coding). 5. Java design pattern for modular implementation of Bio. Moby web services that get their data from any GCP-compliant Data. Source that supports a given find() use case.

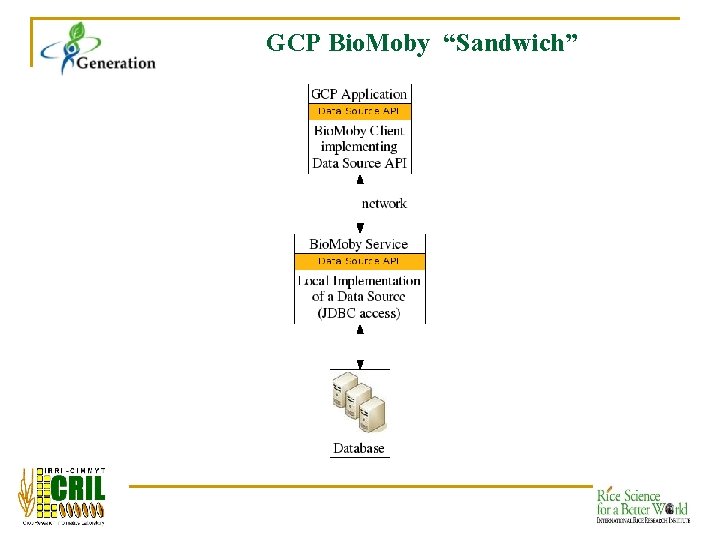
GCP Bio. Moby “Sandwich”

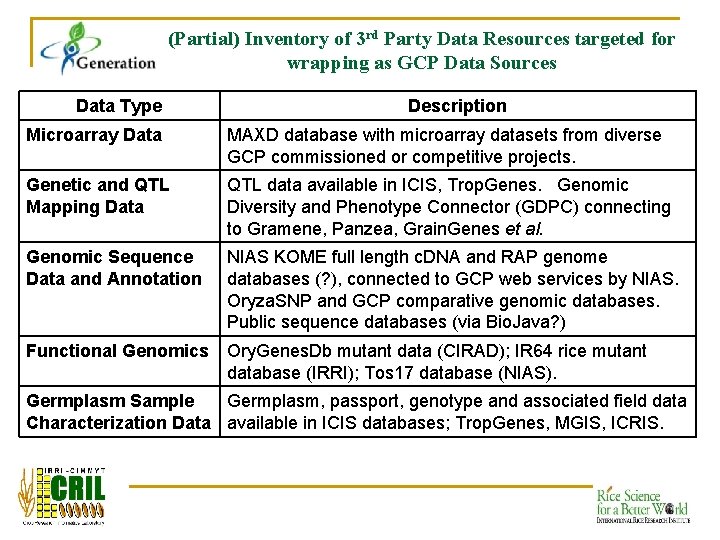
(Partial) Inventory of 3 rd Party Data Resources targeted for wrapping as GCP Data Sources Data Type Description Microarray Data MAXD database with microarray datasets from diverse GCP commissioned or competitive projects. Genetic and QTL Mapping Data QTL data available in ICIS, Trop. Genes. Genomic Diversity and Phenotype Connector (GDPC) connecting to Gramene, Panzea, Grain. Genes et al. Genomic Sequence Data and Annotation NIAS KOME full length c. DNA and RAP genome databases (? ), connected to GCP web services by NIAS. Oryza. SNP and GCP comparative genomic databases. Public sequence databases (via Bio. Java? ) Functional Genomics Ory. Genes. Db mutant data (CIRAD); IR 64 rice mutant database (IRRI); Tos 17 database (NIAS). Germplasm Sample Germplasm, passport, genotype and associated field data Characterization Data available in ICIS databases; Trop. Genes, MGIS, ICRIS.

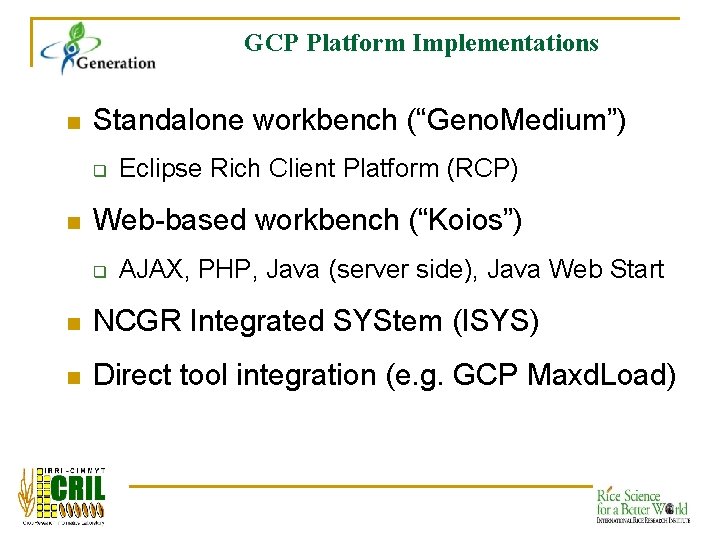
GCP Platform Implementations n Standalone workbench (“Geno. Medium”) q n Eclipse Rich Client Platform (RCP) Web-based workbench (“Koios”) q AJAX, PHP, Java (server side), Java Web Start n NCGR Integrated SYStem (ISYS) n Direct tool integration (e. g. GCP Maxd. Load)

http: //moby. generationcp. org

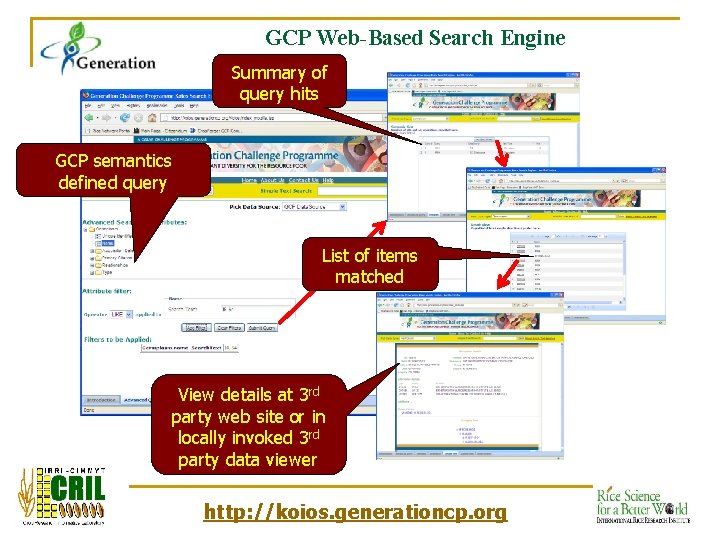
GCP Web-Based Search Engine Summary of query hits GCP semantics defined query List of items matched View details at 3 rd party web site or in locally invoked 3 rd party data viewer http: //koios. generationcp. org

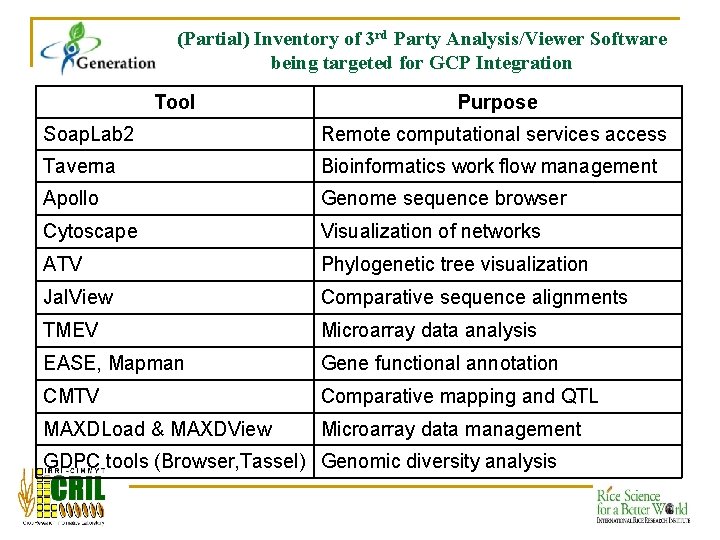
(Partial) Inventory of 3 rd Party Analysis/Viewer Software being targeted for GCP Integration Tool Purpose Soap. Lab 2 Remote computational services access Taverna Bioinformatics work flow management Apollo Genome sequence browser Cytoscape Visualization of networks ATV Phylogenetic tree visualization Jal. View Comparative sequence alignments TMEV Microarray data analysis EASE, Mapman Gene functional annotation CMTV Comparative mapping and QTL MAXDLoad & MAXDView Microarray data management GDPC tools (Browser, Tassel) Genomic diversity analysis

GCP “Pantheon” Project in Crop. Forge http: //cropforge. org/projects/pantheon/

Closing Perspective n The GCP is a global consortium of 22++ crop research partners who need to share diverse large data sets and tools, in a globally distributed manner. n Given the scope and duration of the GCP, developers within the consortium embraced the task of developing public global informatics standards for interoperability and integration. n The effort is an open source, global community building exercise. n We welcome the participation of any and all interested scientists and developers who might wish to use and/or contribute to the further evolution and application of these standards.