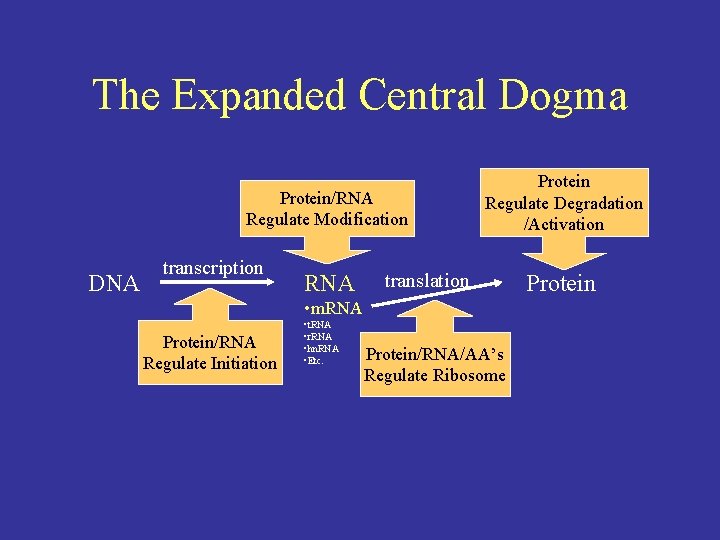
The Expanded Central Dogma ProteinRNA Regulate Modification DNA
- Slides: 8

The Expanded Central Dogma Protein/RNA Regulate Modification DNA transcription RNA Protein Regulate Degradation /Activation translation • m. RNA Protein/RNA Regulate Initiation • t. RNA • r. RNA • hn. RNA • Etc. Protein/RNA/AA’s Regulate Ribosome Protein

c. DNA Microarrays • Takes “snapshot” of cellular m. RNA levels under chosen experimental conditions • Two current representations: – General un-weighted graph (enumerate maximal cliques, neighborhood search, dominating set) – Bipartite graph (biclique) • Goal: Group genes that exhibit similar responses to stimuli – co-regulation

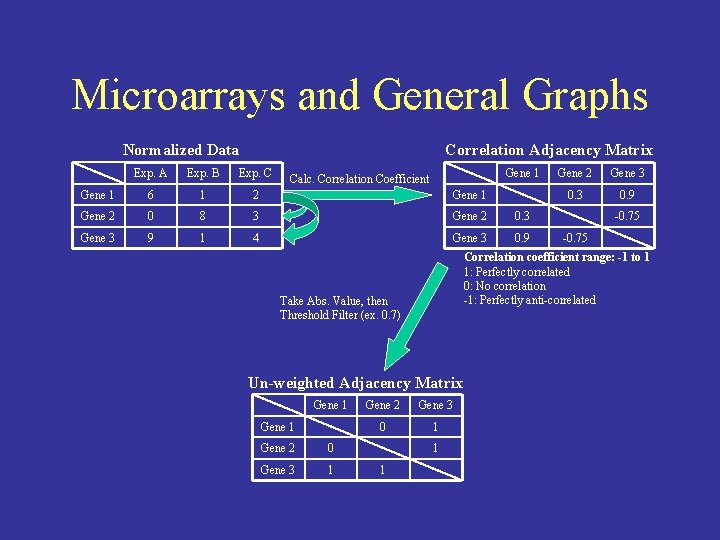
Microarrays and General Graphs Normalized Data Correlation Adjacency Matrix Exp. A Exp. B Exp. C Gene 1 6 1 2 Gene 1 Gene 2 0 8 3 Gene 2 0. 3 Gene 3 9 1 4 Gene 3 0. 9 Calc. Correlation Coefficient Un-weighted Adjacency Matrix Gene 1 Gene 2 0 Gene 3 1 Gene 2 Gene 3 0 1 1 1 Gene 3 0. 9 -0. 75 Correlation coefficient range: -1 to 1 1: Perfectly correlated 0: No correlation -1: Perfectly anti-correlated Take Abs. Value, then Threshold Filter (ex. 0. 7) Gene 1 Gene 2

Clique Enumeration Algorithm • Choose ‘well-selected’ vertex (no pre-processing) • Add selected vertex to set compsub (potential clique) • Create sets candidate and not from old sets by removing all vertices not connected to the selected candidate (keeping old intact) • Call function recursively on new sets • On return, remove selected vertex from set compsub and add to set not • Clique is found when sets candidate and not are empty

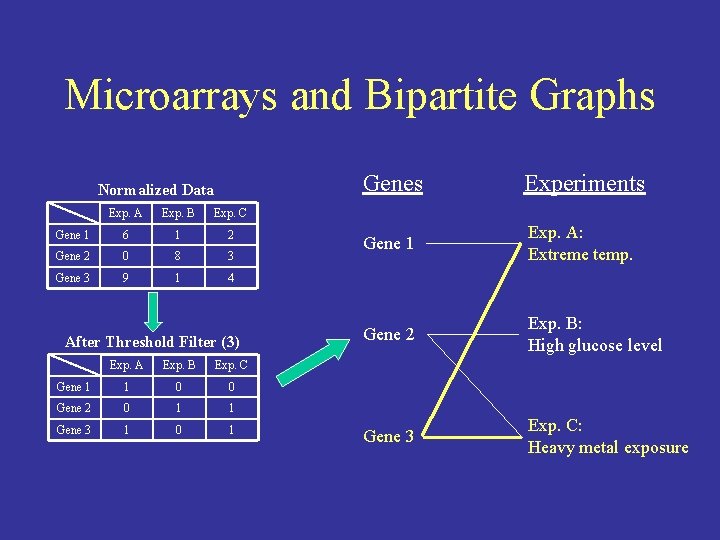
Microarrays and Bipartite Graphs Normalized Data Exp. A Exp. B Exp. C Gene 1 6 1 2 Gene 2 0 8 3 Gene 3 9 1 4 After Threshold Filter (3) Exp. A Exp. B Exp. C Gene 1 1 0 0 Gene 2 0 1 1 Gene 3 1 0 1 Genes Experiments Gene 1 Exp. A: Extreme temp. Gene 2 Exp. B: High glucose level Gene 3 Exp. C: Heavy metal exposure

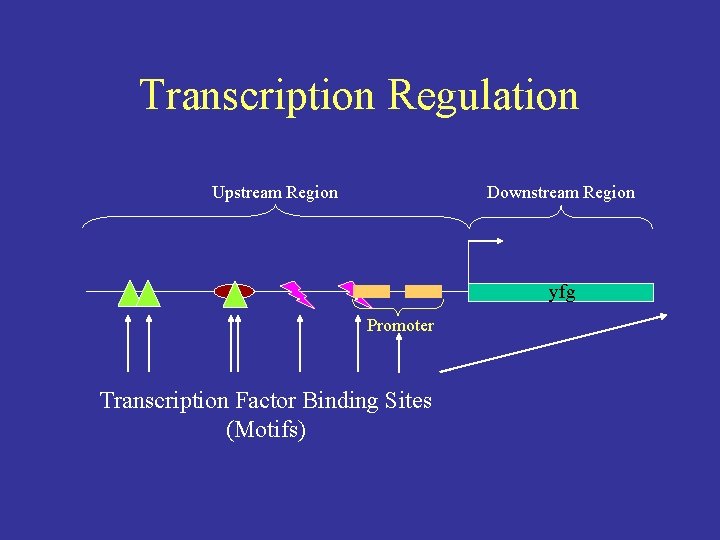
Transcription Regulation Upstream Region Downstream Region yfg Promoter Transcription Factor Binding Sites (Motifs)

Module Detection • Presence of common motifs indicates coordinate regulation – a module • Sequence of motifs in module rarely important • Position of a motif relative to other motifs can be important

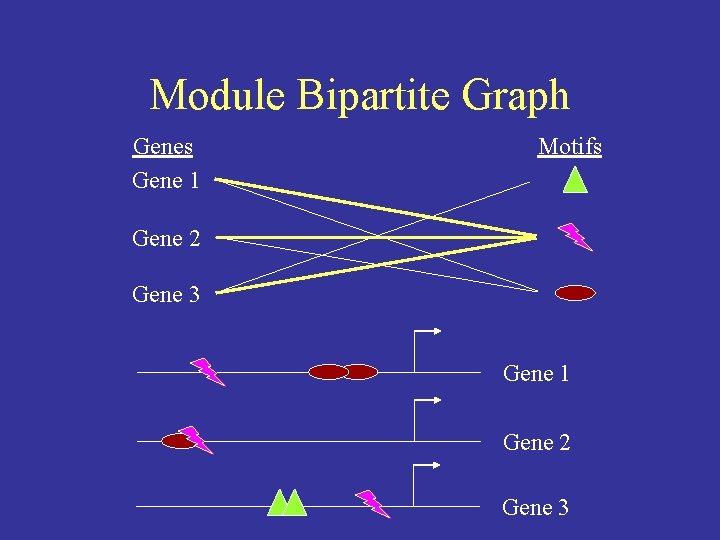
Module Bipartite Graph Genes Gene 1 Motifs Gene 2 Gene 3 Gene 1 Gene 2 Gene 3